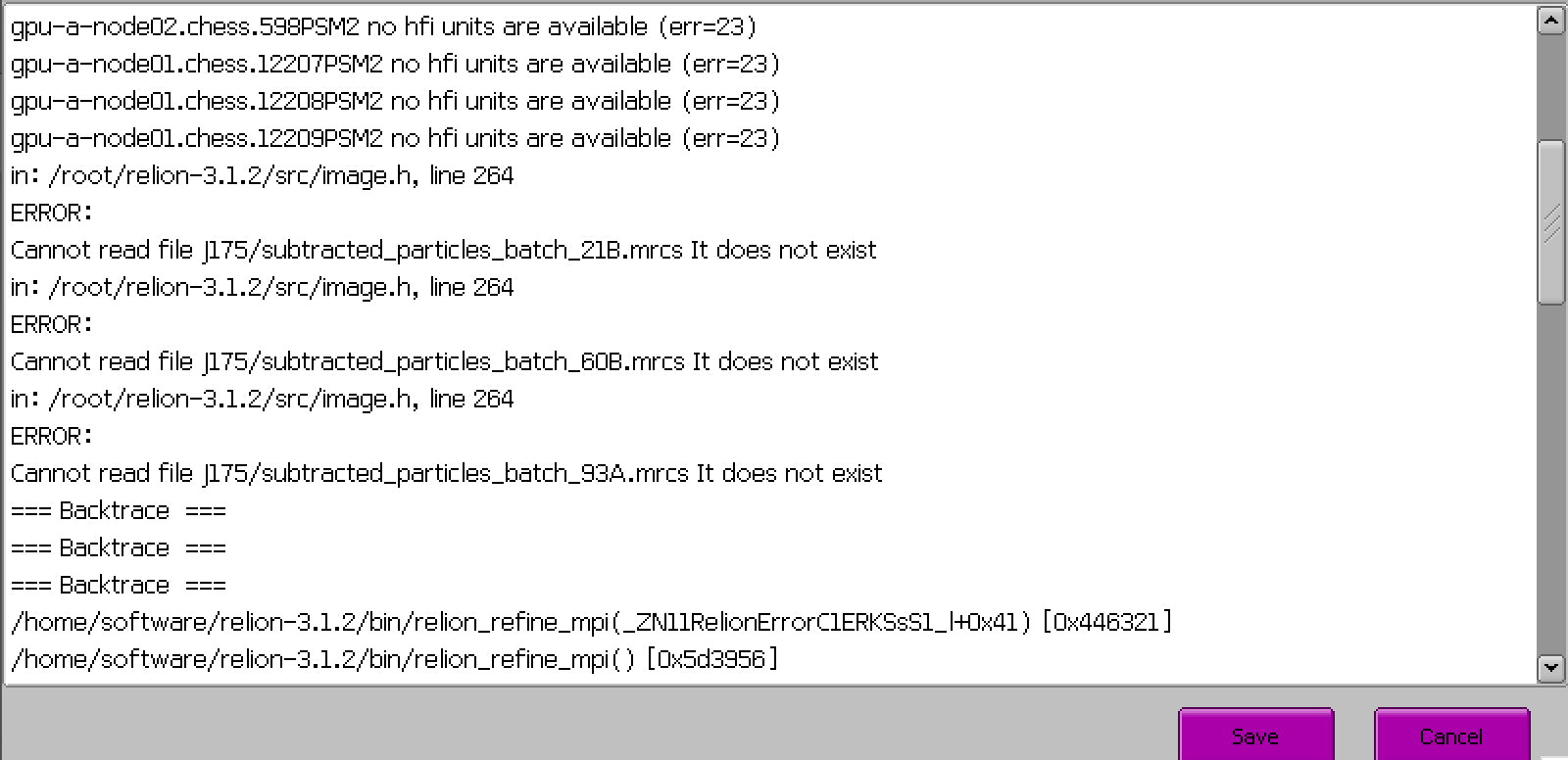
Hi, everyone. I generate a star file by using Pyem. But I can’t use it run 3d classification successfully in relion. Why? The error is shown below.
The process:
csparc2star.py cryosparc_P10_J191_009_particles.cs from_csparc.star
J191 is local refinement, its particle input from J175 - particle subtraction.
In relion(3d classification):
Input images STAR file: from_csparc.star
Reference map: J191_003_volume_map.mrc
have you linked the J175 dir to your relion dir? otherwise it won’t work
@olibclarke Thank you.
So I should do like this, right? But the directory should be make in what?
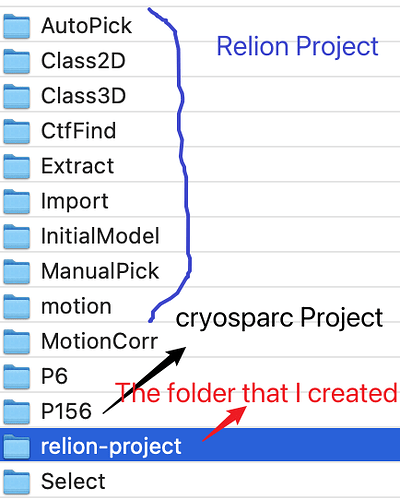
I generated .star file on local, then uploaded our cluster and linked the 175 dir to relion-project. I am not sure what I did is not correct. In another words, the relion-project should be which one. I just created a folder in the same directory as the Relion. See the picture below:
Look at the star file in a text editor. The relative paths that it is looking for have to exist, otherwise you will get errors like you encountered. Make soft links so the files are where the star file expects them to be
Only this information in the Star file. Should I make soft links with J120?
data_optics
loop_
_rlnVoltage #1
_rlnImagePixelSize #2
_rlnSphericalAberration #3
_rlnAmplitudeContrast #4
_rlnOpticsGroup #5
_rlnImageSize #6
_rlnImageDimensionality #7
300.000000 0.660000 2.700000 0.100000 1 500 2
300.000000 0.660000 2.700000 0.100000 2 500 2
data_particles
loop_
_rlnImageName #1
_rlnMicrographName #2
_rlnCoordinateX #3
_rlnCoordinateY #4
_rlnAngleRot #5
_rlnAngleTilt #6
_rlnAnglePsi #7
_rlnOriginXAngst #8
_rlnOriginYAngst #9
_rlnDefocusU #10
_rlnDefocusV #11
_rlnDefocusAngle #12
_rlnPhaseShift #13
_rlnCtfBfactor #14
_rlnClassNumber #15
_rlnOpticsGroup #16
000002@J175/subtracted_particles_batch_0A.mrcs J120/motioncorrected/20220316_jlt_0001_patch_aligned_doseweighted.mrc 3751 1019 -110.801620 75.430527 -170.485580 0.346500 1.633500 12391.720703 11660.912109 -73.842682 0.000000 0.000000 4 2
No, you need to soft link the J175 dir to your relion dir
OK. I understand. Thank you very much.