Hello All. I’m a bit uncertain of how to approach 3D refinements of a particular protein. I’m working with a large, tetrameric channel that exhibits some large motion in its cytoplasmic domains. I don’t think what I have is an uncommon problem, but I’m uncertain of how to proceed. I’ll start from the top:
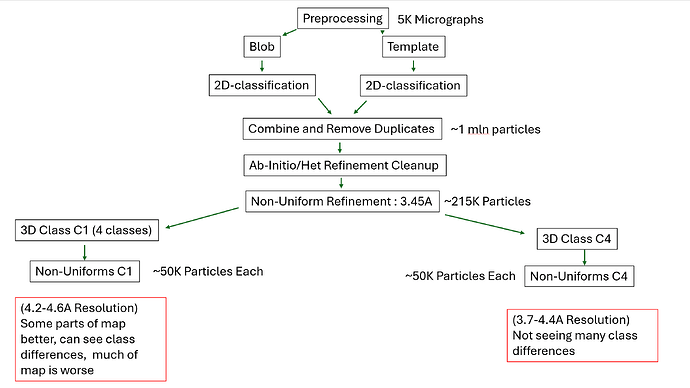
After picking I’m left with a million particles. Using C4 ab-initios and hetero-refinements with decoys I have a clean particle stack of ~215K particles. A C4 non-uniform refinement gives me a 3.45A map with an OK TMD in C4, but the map is deficient at the cytoplasmic domains.
Essentially, I am presented with a region of my protein that’s in C4 but has four, individual domains which are moving around a bit that are not in any symmetry. I want to see if I can separate these classes out, which has been done before for this protein but the documentation is lacking.
This is where I could be overthinking things. I want to 3D class this protein and then try non-uniform and local refinements of just one cytoplasmic domain to see if I can A. separate discrete classes and B. improve the maps. But how should I apply symmetry?
I’ve done both 3D classification in C1 and C4 and refined the outputs. The C1 outputs are worse in terms of resolution and much of the map, but I can see much more motion (as I expect) of the cytoplasmic domains.
The 3D classification for C4 gives me some slightly worse resolutions (I suspect I need more particles based off of reslog), but the maps still have deficiencies in the cytoplasmic domains.
I was going to proceed with trying some local refinements on the C1 maps (masks and refinements both for TMD and cytoplasmic domains) but I have a suspicion that I am missing something here.