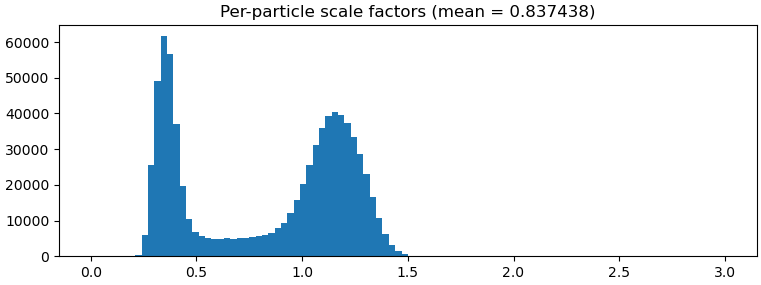
I occasionally have binomial/multi-modal distribution in my per-particle scale factor histogram and would like to slice and reconstruct the different populations of particles to figure out what is going on. Could you please implement this action in the GUI. Perhaps in a new tool that also includes other ways of splitting the data, e.g. based on defocus value, X and Y coordinates etc. It could be an interactive job akin to the manual curate exposure or inspect particle picks.
Thanks @daniel.s.d.larsson for your suggestion. I realize that you specifically requested a GUI implementation, but wanted to mention that cryosparc-tools might be a good fit for this task.
@wtempel, how would you do the selection in cryosparc-tools?
I have my particles dataset loaded and can see the alignments3D/alpha column, which seems to be the per-particle scale factors, but I’m not sure how to split the dataset based on some threshold (eg. keep all >0.6).
Can I use query with a range? Or maybe split_by?
I must say a GUI implementation of this feature would be most welcome
Following – did anyone make any progress on this? I think it would be very useful!
Hi @JacobLewis! Here is an example notebook using cryosparc-tools which will filter based on per-particle scale. The selected scale threshold is set by changing scale_threshold.
I hope that is helpful!
In addition to other suggestions here, you can also do this using 3D Flex data prep - it is a bit of a hack but for a simple bimodal distribution like this it works fine
hi @rwaldo awesome! I’m going to test this out! thanks for taking the time!
Does anyone have results showing the difference between low-scale & high-scale values? I got my data with bi-modal distribution in scale as well.
We often see that the low-scale particles correspond to junk - but it’s always worth checking, you can just split by scale and then run 2D classification to see what is there
Thanks, I will try and report back. I have the same feeling and if it works it is fantastic since in our case, it is not easy to remove junk.
I tried to use cryosparc tools to do it. First time and also no administrative right on it.
I have problem with the:
with open(‘/u/rposert/instance-info.json’, ‘r’) as f:
instance_info = json.load(f)
What should I put in here for my own username?
Thanks.
I would just use 3D-Flex data prep
My god, the 3D-Flex data prep gonna take forever since I got ~1M particles at 400px ![]()
Shouldn’t take that long? I did 260k particles at 450px and it took 18min…
Perhaps it is a cluster with 2080 Ti. The 3090 node is not available today and data transfer might be the bottle neck. 2.5hr and only 1/3 of the particles.
H
Ok, I managed to get the cryosparc tools work.
The json file is basically this:
cs = CryoSPARC(
license=“xxxxxxxx-xxxx-xxxx-xxxx-xxxxxxxxxxxx”,
host=“localhost”,
base_port=39000,
email="ali@example.com",
password=“password123”
)
email & password is for your account only, no need the admin password.
Hmm @rwaldo
I got this error when writing out the particles set. Can you help me? Just my data is filament data.
Error in Jupyter Notebook
/data/xxx/.conda/envs/cryosparctools/lib/python3.10/contextlib.py:135: UserWarning: *** CommandClient: (http://localhost:61003/external/projects/P6/jobs/J761/outputs/particle/dataset) HTTP Error 422 UNPROCESSABLE ENTITY; please check cryosparcm log command_vis for additional information.
Response from server: b"Invalid dataset; missing the following required fields: {(‘filament/start_frac’, ‘<f4’, (2,)), (‘filament/filament_uid’, ‘<u8’), (‘filament/straight_length_A’, ‘<f4’), (‘filament/position_A’, ‘<f4’), (‘filament/end_frac’, ‘<f4’, (2,)), (‘filament/curvature’, ‘<f4’), (‘filament/sinuosity’, ‘<f4’), (‘filament/inter_box_dist_A’, ‘<f4’), (‘filament/arc_length_A’, ‘<f4’)}"
Update: I modified the code a bit, add the "slots=[“alignments3D”], line and it worked. Don’t know if I do the right thing or not. Seems to be correct.
cs.save_external_result(
project_number,
workspace_number,
filtered_particles,
slots=[“alignments3D”],
type = ‘particle’,
passthrough = (job_number, ‘particles’),
title = f"Filtered per-particle scale <= {scale_threshold}"
)
Update 2: I just want to update here that the low-scale particles are still mostly correct particles in my case but seem to have lower quality. 2D class max resolution from 200k particles is only at a maximum 9 Angstrom from a 3.3 Angstrom structure.
Hi @builab — it’s a good point that I never really explain what instance-info.json is. You’re right that you can replace it with the “normal” way of authenticating the CryoSPARC() object. I’ve added a note on what the JSON file is to the examples README. Thanks for the note!
As far as your notebook error, I’m not sure why you need to include the aligments3D from the filtered dataset, but I’m glad you got it working!