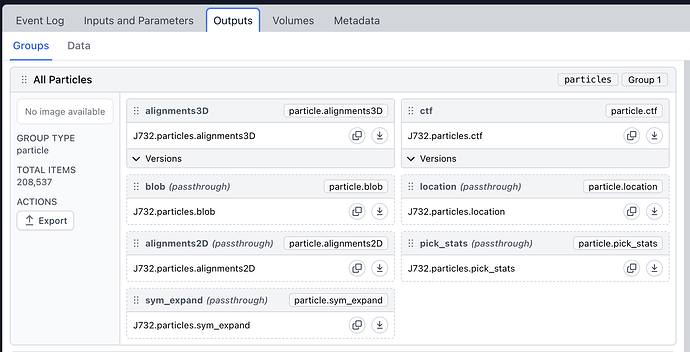
Tried this, but when I run it I get this error:
*** CommandClient: (http://localhost:39003/external/projects/P48/jobs/J745/outputs/particle/dataset) HTTP Error 422 UNPROCESSABLE ENTITY; please check cryosparcm log command_vis for additional information.
Response from server: b'Invalid dataset stream'
Traceback (most recent call last):
File "split_by_symm.py", line 35, in <module>
project.save_external_result(
File "/home/user/.local/lib/python3.8/site-packages/cryosparc/project.py", line 278, in save_external_result
return self.cs.save_external_result(
File "/home/user/.local/lib/python3.8/site-packages/cryosparc/tools.py", line 501, in save_external_result
job.save_output(output, dataset)
File "/home/user/.local/lib/python3.8/site-packages/cryosparc/job.py", line 1323, in save_output
with make_request(self.cs.vis, url=url, data=dataset.stream()) as res:
File "/home/user/software/miniconda3/envs/cryosparc-tools/lib/python3.8/contextlib.py", line 113, in __enter__
return next(self.gen)
File "/home/user/.local/lib/python3.8/site-packages/cryosparc/command.py", line 192, in make_request
raise CommandClient.Error(client, error_reason, url=url)
cryosparc.command.Error: *** CommandClient: (http://localhost:39003/external/projects/P48/jobs/J745/outputs/particle/dataset) HTTP Error 422 UNPROCESSABLE ENTITY; please check cryosparcm log command_vis for additional information.
Response from server: b'Invalid dataset stream'
When I just print(filtered_dataset) the output looks sensible, so I think something is going wrong in the saving of external results - this is using Python 3.8.19, and CS4.4.0
log of command_vis:
2024-03-25 16:00:47,347 request_handler INFO | Received request for get_project_file
2024-03-25 16:00:47,384 request_handler INFO | Completed request for get_project_file in 0.04s
2024-03-25 16:00:48,215 request_handler INFO | Received request for get_project_file
2024-03-25 16:00:48,258 request_handler INFO | Completed request for get_project_file in 0.04s
2024-03-25 16:00:52,565 upload_external_job_output INFO | Received external job output P48.J747.particle
2024-03-25 16:00:52,762 upload_external_job_output ERROR | Invalid dataset stream
2024-03-25 16:00:52,762 upload_external_job_output ERROR | Traceback (most recent call last):
2024-03-25 16:00:52,762 upload_external_job_output ERROR | File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_tools/cryosparc/dataset.py", line 559, in load
2024-03-25 16:00:52,762 upload_external_job_output ERROR | raise TypeError(f"Could not determine dataset format (prefix is {prefix})")
2024-03-25 16:00:52,762 upload_external_job_output ERROR | TypeError: Could not determine dataset format (prefix is b'\x94CSDAT')
2024-03-25 16:00:52,762 upload_external_job_output ERROR |
2024-03-25 16:00:52,762 upload_external_job_output ERROR | The above exception was the direct cause of the following exception:
2024-03-25 16:00:52,762 upload_external_job_output ERROR |
2024-03-25 16:00:52,762 upload_external_job_output ERROR | Traceback (most recent call last):
2024-03-25 16:00:52,762 upload_external_job_output ERROR | File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_command/command_vis/snowflake.py", line 395, in upload_external_job_output
2024-03-25 16:00:52,762 upload_external_job_output ERROR | dset = Dataset.load(request.stream)
2024-03-25 16:00:52,762 upload_external_job_output ERROR | File "/home/user/software/cryosparc/cryosparc2_master/cryosparc_tools/cryosparc/dataset.py", line 592, in load
2024-03-25 16:00:52,762 upload_external_job_output ERROR | raise DatasetLoadError(f"Could not load dataset from file {file}") from err
2024-03-25 16:00:52,762 upload_external_job_output ERROR | cryosparc_tools.cryosparc.errors.DatasetLoadError: Could not load dataset from file <werkzeug.serving.DechunkedInput object at 0x7fdbc5e6ecd0>
EDIT: Figured out the issue - when csparc tool was installed (this morning) as per the guide, I installed it with pip install cryosparc-tools. However, it turns out this installs v4.3, not v4.4. When I install it with pip install cryosparc-tools~=4.4.0, the script now works without a problem. I wonder if it would be worth having an automatic check somewhere for a version mismatch, so this is flagged earlier and in a more obvious way?