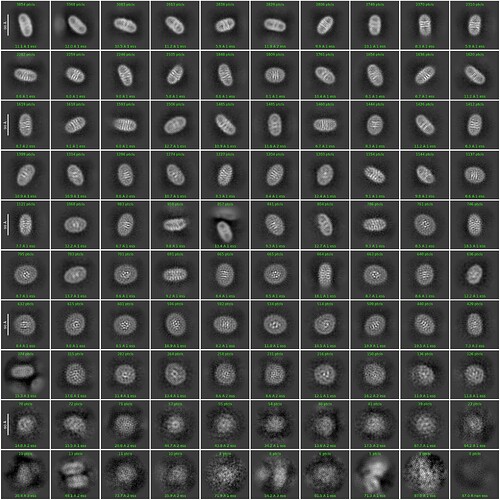
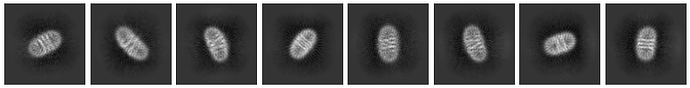
Hello, everyone. I’m new to the field of cryo-electron microscopy and currently working on a small protein of around 60 kDa. I have reconstituted it into nanodiscs and collected about 3,000 micrographs. I have got some not bad 2D classification result, but can not reconstruct the ab-initio model that could be used for further processing. Could anyone share advice or suggest solutions to help overcome this issue? Thank you!
This is my parameters:
2d classification:
Number of 2D classes :100
Number of online-EM iterations: 40
Batchsize per class: 200
I have tried to turn off Force max over poses/shifts, but can not help me to generate the excellent result. Should I try other parameters?
Ab-initio:
Number of Ab-Initio classes: 3
Maximum resolution (Angstroms): 6-8
Initial resolution (Angstroms) :7-15
Class similarity:0
Actually, I realized that mderating batchsize could help to class better in ab-initio job, and I have attempted but not work.
If anyone can give me some advice, I would be grateful, thanks again!