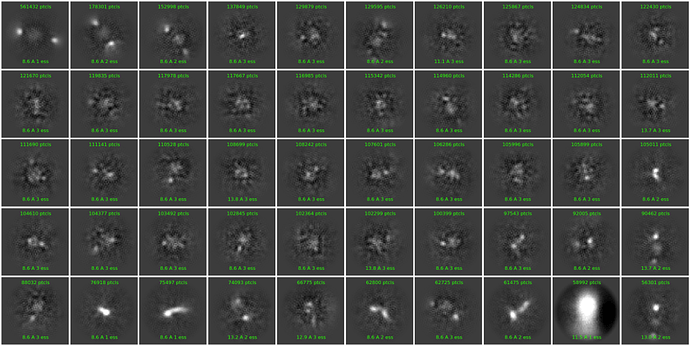
Hi, I’m new here and trying to analyze a relatively small protein/RNA complex with a total molecular weight of around 92.7 kDa. The issue is that after the initial grid scan, it seemed we obtained potentially usable data, so we proceeded to collect it. However, we ended up with very blurry 2D images that look like noise or a ton of junk particles. I can’t figure out why a seemingly good result after scanning could lead to such a mess in the actual analysis process. No doubt we failed to construct a good 3D model…
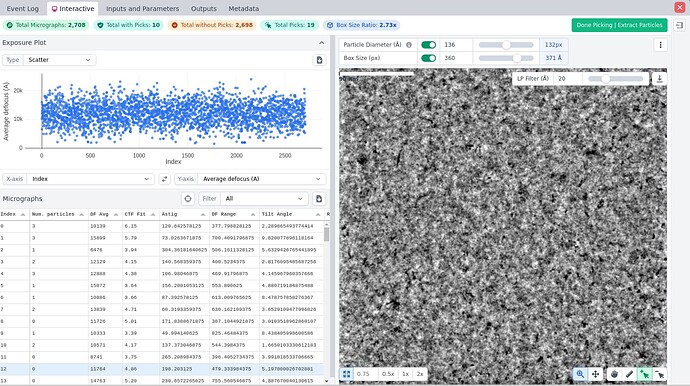
Below are some pictures of my sample grid square and the confusing 2D images. Could anyone kindly assist me in explaining this puzzling situation or offer me some advice for improvement?
-
What does your picking look like?
-
There seems to be an issue with your gain correction, you may want to try making a gain reference from your movies (e.g. using
relion_estimate_gain)
that is an insanely high number of particles per class. make sure your picking is faithful to each 1 particle. perhaps manual pick 100, then generate 3 2D classes and then template pick. if you just have an insanely high number of micrographs, then you might consider processing 5000 micrographs at a time. your micrographs have strong evidence for ~200-400 particles each.
Thank you for your response. It appears that we have already completed the gain reference process at the beginning of the data input. Here is one template pick image. Unfortunately, I can’t discern much from it. ![]()
Thanks for your reply. It sounds like a practical solution, and I will give it a try. However, I am facing challenges with my samples, making it difficult to discern potential particles from the background. Here is a snapshot representing the typical condition.
As suggested by @CryoEM2 - you are way overpicking. In your initial images, particles can actually be discerned, but probably only 50-100 per mic I would say.
Re the gain - I was not suggesting that you had not done gain correction, but there are consistent patterns in your initial images which suggest the gain reference is not optimally matched to your data - something to investigate.
In cases where particles are challenging to identify, you might consider training a denoising model for your data, using the new denoising job.
Looks like a good start. Load result to “inspect particle picks” job, slide the NCC score and Power score thresholds until there are only 100 picks per image (if feeling confident, adjust them until you faithfully pick all particles and not much background). Extract and run 2D again.
The particle picker jobs all do a great job of 1) finding the good hits that are obvious 2) finding lots of extra stuff that “might” be a particle. It’s up to you to take the result and limit the output only to the appropriate picks.