Hi CryoSPARC community,
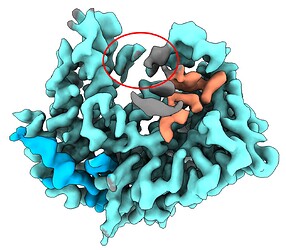
I am currently processing a small molecule (~60 kDa), where I suspect a single-stranded RNA might be buried in the groove, though the sequence is uncertain. In my final density map (300k particles, 3.2 Å), I’ve managed to model the protein, and most atoms are assigned (represented in blue and light blue). However, there are regions where density is unaccounted for — the grey density could be from subtracted or flexible RNA. I’ve modeled four RNA molecules into the light salmon density, and there is also density circled in red ellipses that seems to have base-like features. Unfortunately, this density is discontinuous.
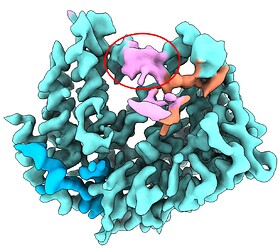
I performed 3D classification (300k particles, 10 classes) with a mask around the RNA density region (dilation radius 4, padding width 12). Refining ~30k good particles using homogeneous-only refinement has resulted in a more continuous density (shown in pink), but the base features are missing. Other classes showed even worse density.
Does anyone have suggestions on how I could further improve the RNA density quality? I’d greatly appreciate any advice.
Best regards,
Yufeng