Hi @yoshiokc. Yes, the motion/path field contains paths to a numpy array, which itself has the rigid + RBMC trajectories. To get the RBMC-only path, you’ll have to load and subtract the RBMC path from this total path. Note also that in the CryoSPARC GUI (but not the data you download here), these trajectories are scaled by a factor of 40 so that they’re visible on the micrograph scale.
Here’s a cryosparc-tools script to help you do this:
from cryosparc.tools import CryoSPARC
import json
import numpy as np
from pathlib import Path
import matplotlib.pyplot as plt
with open(Path('~/instance-info.json').expanduser(), 'r') as f:
instance_info = json.load(f)
cs = CryoSPARC(**instance_info)
assert cs.test_connection()
project_uid = "P312"
job_uid = "J142"
project = cs.find_project(project_uid)
rbmc = project.find_job(job_uid)
particles_rbmc = rbmc.load_output("particles_0")
pmc_juids = np.unique([x.split('/')[0] for x in particles_rbmc["location/micrograph_path"]])
pmc_micrographs = {puid: project.find_job(puid).load_output("micrographs") for puid in pmc_juids}
downloaded_motion_files = {}
def get_trajectories(particle_uid):
p_rbmc = particles_rbmc.query({"uid": particle_uid})[0]
total_motion_filename = p_rbmc["motion/path"]
if total_motion_filename not in downloaded_motion_files:
downloaded_motion_files[total_motion_filename] = np.load(project.download_file(total_motion_filename))
total_trajectory = downloaded_motion_files[total_motion_filename][p_rbmc["motion/idx"]]
mic_uid = p_rbmc["location/micrograph_uid"]
pmc_juid = p_rbmc["location/micrograph_path"].split("/")[0]
pmc_mic = pmc_micrographs[pmc_juid].query({"uid": mic_uid})[0]
rigid_motion_filename = pmc_mic["rigid_motion/path"]
if rigid_motion_filename not in downloaded_motion_files:
downloaded_motion_files[rigid_motion_filename] = np.load(project.download_file(rigid_motion_filename))
rigid_trajectory = np.squeeze(downloaded_motion_files[rigid_motion_filename])
rbmc_trajectory = total_trajectory - rigid_trajectory
return total_trajectory, rigid_trajectory, rbmc_trajectory
So, for example:
uid = 14502624203646355101
total, rigid, rbmc = get_trajectories(uid)
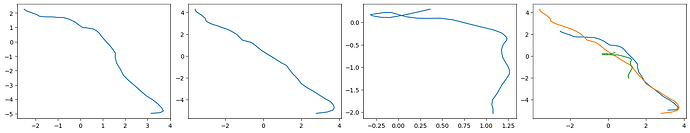
fig, axs = plt.subplots(1, 4, figsize = (16, 3), frameon = False, layout = "constrained")
# I have no idea why the forum is coloring these blue??
axs[0].plot(total[:,0], total[:,1])
axs[3].plot(total[:,0], total[:,1])
axs[1].plot(rigid[:,0], rigid[:,1])
axs[3].plot(rigid[:,0], rigid[:,1])
axs[2].plot(rbmc[:,0], rbmc[:,1])
axs[3].plot(rbmc[:,0], rbmc[:,1])
Makes this plot: