I have done the initial processing in CryoSPARC (including patch-motion corection and patch ctf) and want to polish those particles in RELION. I’ve exported the CryoSPARC particles and converted them using csparc2star.py including the --inverty option. In RELION the movies were processed and are motioncorrected and ctf estimated, from those I now want to re-extract.
I’ve adjusted the micrograph path in the particle.star file (interestingly Relion changed every . in the Micrograph in the name to _ ) and made sure the opticsGroup is matching.
My particle.star file looks like this:
# version 30001
data_optics
loop_
_rlnVoltage #1
_rlnImagePixelSize #2
_rlnSphericalAberration #3
_rlnAmplitudeContrast #4
_rlnOpticsGroup #5
_rlnImageSize #6
_rlnImageDimensionality #7
_rlnOpticsGroupName #8
300.000000 1.050000 2.700000 0.100000 1 384 2 opticsGroup1
# version 30001
data_particles
loop_
_rlnImageName #1
_rlnMicrographName #2
_rlnCoordinateX #3
_rlnCoordinateY #4
_rlnAngleRot #5
_rlnAngleTilt #6
_rlnAnglePsi #7
_rlnOriginXAngst #8
_rlnOriginYAngst #9
_rlnDefocusU #10
_rlnDefocusV #11
_rlnDefocusAngle #12
_rlnPhaseShift #13
_rlnCtfBfactor #14
_rlnOpticsGroup #15
_rlnRandomSubset #16
_rlnClassNumber #17
_rlnGroupNumber #18
_rlnNormCorrection #19
_rlnLogLikeliContribution #20
_rlnMaxValueProbDistribution #21
_rlnNrOfSignificantSamples #22
_rlnImageOriginalName #23
000001@J34/extract/000002298445352262704_2024-05-13_20.42.57_40S-1v2_87-5_00012_30.0_1-12_patch_aligned_doseweighted_particles.mrcs MotionCorr/job015/frames/2024-05-13_20_42_57_40S-1v2_87-5_00012_30_0_1-12.mrc 1426.000000 1985.000000 115.120843 69.853913 99.233130 12.431996 13.481996 12339.794922 12281.777344 264.055359 0.000000 0.000000 1 2 1 1 0.606181 6.187094e+05 0.457129 5 000001@Subtract/job003/Particles/subtracted_rank1_opticsgroup1.mrcs
and my micrograph file looks like this:
# version 30001
data_optics
loop_
_rlnOpticsGroupName #1
_rlnOpticsGroup #2
_rlnMicrographOriginalPixelSize #3
_rlnVoltage #4
_rlnSphericalAberration #5
_rlnAmplitudeContrast #6
_rlnMicrographPixelSize #7
opticsGroup1 1 1.050000 300.000000 2.700000 0.100000 1.050000
# version 30001
data_micrographs
loop_
_rlnCtfPowerSpectrum #1
_rlnMicrographName #2
_rlnMicrographMetadata #3
_rlnOpticsGroup #4
_rlnAccumMotionTotal #5
_rlnAccumMotionEarly #6
_rlnAccumMotionLate #7
MotionCorr/job015/frames/2024-05-13_15_57_27_40S-1v2_86-1_00001_30_0_1-1_PS.mrc MotionCorr/job015/frames/2024-05-13_15_57_27_40S-1v2_86-1_00001_30_0_1-1.mrc MotionCorr/job015/frames/2024-05-13_15_57_27_40S-1v2_86-1_00001_30_0_1-1.star 1 42.934522 18.514962 24.419560
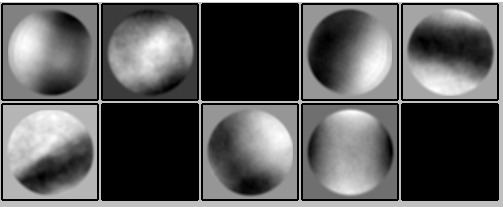
However, when I extract my particles and run a 2D classification to confirm I don’t get any particles:
My initial thought was that something has to be off with the coordinate system, therefore I compared the coordinates from an ice crystal between a CryoSPARC processed micrograph and a Relion processed micrograph and the are the same (when taking in account that from CryoSPARC to Relion the Y-axis needs to be inverted)
Do you know what could’ve gone wrong during conversion? I tried the same protocol on a different dataset and there it worked (different microscope and no classification in Relion before tring to re-extract).
Thanks!
How did you compare them?
There must be some difference that slipped past then? You probably need --flipy as well btw, as the micrographs are upside down in Relion vs. cryoSPARC motion correction (in addition to the convention flip).
Are you using the latest pyem and your steps look like Method 0 from pyem wiki?
Thank you for the suggestions, and sorry for the late reply!
How did you compare them?
I downloaded the same micrograph one processed with CryoSPARC and one processed with Relion and opened them with ChimeraX. There I checked the coordinates for multiple distinct features. The x coordinate was the same, the y coordinate was different. After I manually inverted the y coordinate in Relion however, it was also the same value.
There must be some difference that slipped past then? You probably need --flipy as well btw, as the micrographs are upside down in Relion vs. cryoSPARC motion correction (in addition to the convention flip).
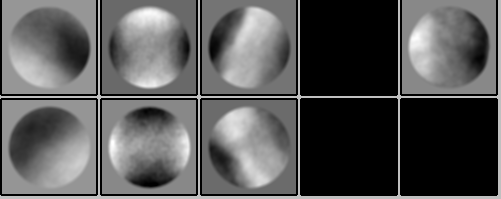
This I just tried (–flipy as an addition to --inverty when running csparc2star.py) and id ended up with no improvement as the 2D classes still look like the particles are extracted in the wrong position:
Are you using the latest pyem
Right now I’m using the pyem version from last october which I should update. Do I need a full re-installation for iit to work? I haven’t bothered with it yet because so far I didn’t have any troubles.
steps look like Method 0 from pyem wiki?
My steps are more or less the same: I exported the particles from CryoSPARC (previously without --flipy but the latest try was with --flipy) and re-extracted them from the Relion MotionCorr Job. The refinement did not work and there I tried 2D classification in order to see what’s wrong with the particles, the result is what you see above. Also I did no use your function --strip-uid and --micrograph-path and just edited them into the particle.star file before re-extracting.
There are some additional dependencies like starfile, otherwise you can just update pyem itself. I don’t remember when various things were fixed last year. Sounds like you have an old version where --inverty doesn’t do anything since you’re saying manually flipping the coordinates works.
Oh sorry, I think there was a misunderstanding: --inverty does work, I just manually did the inversion to see if --inverty gives the correct y-value.
However, I’ll update pyem and give it another go!
Here’s how it should work:
- If you open the same micrograph cryosparc/patch motion and RelionCorr versions, the Relion one should be upside down as compared to cryosparc (or .tif unaligned sum).
- cryosparc coordinate origin in bottom left, normalized coordinates
- Relion coordinate origin in top left, integer pixel coordinates
- ergo when different motion correction is used, and extraction is done in Relion, the two flips cancel (
--inverty)
- note cryosparc flips coordinates on import (i.e. Relion coords are imported correctly), thus
--inverty is also needed to cycle coordinates in and out of cryosparc with unchanged values
- so no invert argument is required when cryosparc Extract Particles and relion Extract are to be used on the same micrographs (original behavior since cryosparc didn’t have movie processing at first)
- the size of the micrograph is taken from the cryosparc metadata not the new micrographs so they will require adjustment if the binning is different
- if the images are upside down but we use the same particle orientations and defocus angles they will be wrong,
--flipy corrects the particle metadata so that relion_reconstruct will work on the converted particles (if you change motion correction both flip metadata and invert coords will be needed!)
- instead one could import the RelionCorr micrographs in cryosparc, run patch motion, import the particles and use Patch Extract to fix the particle CTFs, then extract, refine, and finally export with no invert arg - this is the safest method approach
--flip should not significantly affect 2D classification results
You should always be able to flip back and forth and one way will work…also there is --invertx but I don’t think you can need it unless you had vertical movies and they got rotated a certain way.
Thank you for that suggestion! I’ll try that the next couple of days and hopefully it’ll work!
FYI I did just have a case going from cryosparc to Relion MotionCorr, --flipy alone was the correct choice. (Relion 5).