Hi all cryosparc fans,
I want to sincerely thank you all for your helpful discussions and tips during my recent postdoc.
The protein I worked on (angiotensin-I converting enzyme) was highly dynamic, relatively small, heterogeneous, and heavily N-glycosylated which posed a number of challenges during image processing. It was really tough to learn cryo-EM remotely from scratch during the pandemic so this forum was a lifeline! Thank you very much 
Here is the link to our Open Access publication in The EMBO Journal: Cryo‐EM reveals mechanisms of angiotensin I‐converting enzyme allostery and dimerization | The EMBO Journal (embopress.org)
Briefly, what made it possible to solve the structures of both monomeric and dimeric ACE from a single dataset (where the dimer was a tiny, tiny fraction which we could not separate biochemically) was:
- initial generation of a 7A structure in RELION by template-based autopicking, iterative 2D classification, and iterative 3D classification
- TOPAZ denoising of the micrographs
- training of a TOPAZ picking model using the 7A particles as positive labels and iteratively improving the model by manual curation and retraining (thanks @tbepler @alexjamesnoble )
- a single round of 2D classification of all TOPAZ-picked particles before going straight to 3D ab initio and 3D heterogeneous refine in cryoSPARC
- extensive 3D sorting in cryoSPARC to separate out different conformational states of the monomer, and separate the monomer from dimer
- 3D variability analysis to identify regions of hinging in the monomer and dimer. This guided mask creation for step 7
- local refinement with the non-uniform refine algorithm in cryoSPARC to focus on a particular domain (without subtraction for the monomer but with subtraction for the dimer since dimerization drastically increased the non-interacting domains’ dynamics) @mmclean @olibclarke
- focused 3D classification w/o alignment in RELION to minimize the heterogeneity in the locally-refined map @DanielAsarnow
- further local refinement in cryoSPARC
- it was ESSENTIAL to use very soft masks to avoid overfitting artefacts due to the high N-glycan content. The masks were dilated to include the protein but exclude most of the glycan density, and then padded very softly to extend beyond the glycan density. @emil
3DVA gave us valuable insight into how intradomain allostery could occur between the different parts of this two-domain enzyme. Although the resolution is not atomic, we hope that the insight we gained could guide future studies into allosteric regulation and dimerization-induced intracellular signalling of ACE.
I hope that my work can assist someone on this forum who is struggling with processing of a similar sample 
monomeric ACE
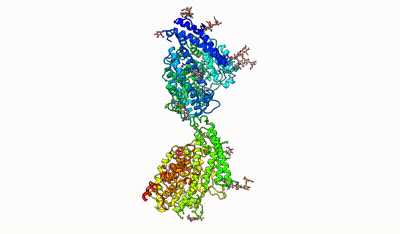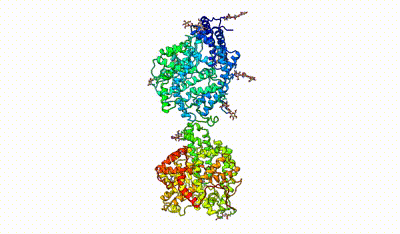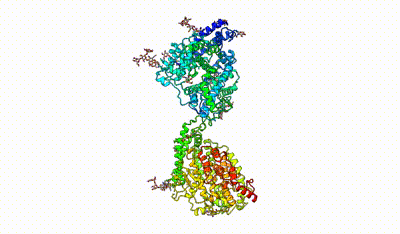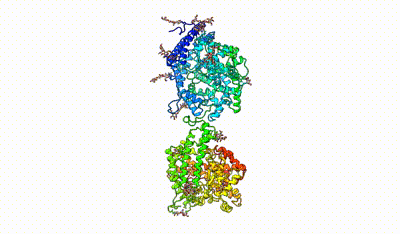
interacting domains of dimeric ACE
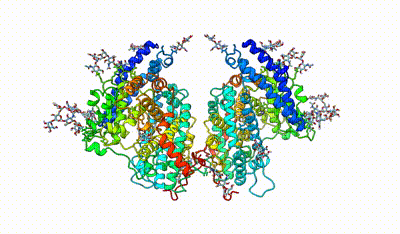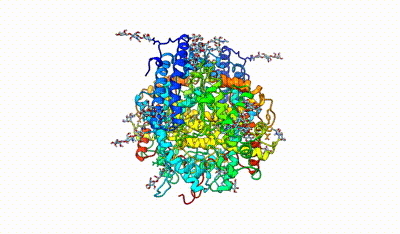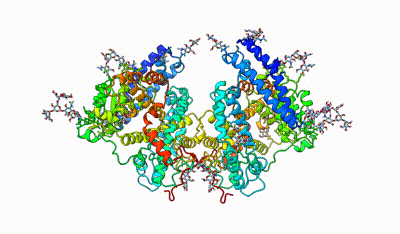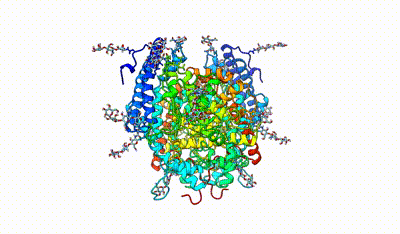
 thank you
thank you