Hello,
I have run into a problem with extracting particles after combining 4 datasets. I believe the error comes from 3 of the datasets being motion-corrected micrographs imported into cryoSPARC, and one dataset being motion corrected within the project.
As some background, we will often import motion corrected micrographs into a new cryoSPARC project and then start the project by running patch CTF estimation. This is done for a variety of reasons, either to prevent duplicating the motion correction job in two projects, the micrographs already being motion corrected in Relion, or if we are bringing previously processed data back onto the local machine.
For these 4 datasets, I processed them in parallel in the same project in separate workspaces. Once I had a clean particle stack for each, I combined the particles for ab initio and 3D refinement, followed by some classification, ultimately getting a 3.4 Ang structure. I now want to try and push the resolution, so I attempted to re-extract and un-bin the particles.
When I ran an extract job (inputs: 4 curate micrographs jobs and the refined particle coordinates from a NU refinement run), I got the following error. One of the datasets would not re-extract. This was the dataset that had been motion corrected within the project. Looking at previous posts, this is related to the background_blob parameter.
Error occurred while processing micrograph J484/motioncorrected/015510707208051555134_n22dec05c_box5g2_00016gr_00039sq940_v02_00005hl_00047esn.frames_patch_aligned_doseweighted.mrc Traceback (most recent call last): File "/data/software/cryosparc/cryosparc2_worker/cryosparc_compute/jobs/pipeline.py", line 61, in exec return self.process(item) File "/data/software/cryosparc/cryosparc2_worker/cryosparc_compute/jobs/extract/run.py", line 488, in process path_abs = os.path.join(proj_dir_abs, micrograph['background_blob/path'])
KeyError: 'background_blob/path'
Marking J484/motioncorrected/015510707208051555134_n22dec05c_box5g2_00016gr_00039sq940_v02_00005hl_00047esn.frames_patch_aligned_doseweighted.mrc as incomplete and continuing...
First, I have double checked and the background_blob passthrough file exists for the problem dataset and is correctly linked to the extract job. However, strangely, it looks like the other three datasets do not have a background_blob entry and extract just fine in this combined extraction job. Furthermore, this dataset will extract fine when the micrograph input is a single curate exposures job with the combined particle coordinates from the NU refine job. Thus, it seems that the KeyError: ‘background_blob/path’ issue only arises when combining data where some micrographs come from a motioncor job within cryosparc and some come from imported aligned micros.
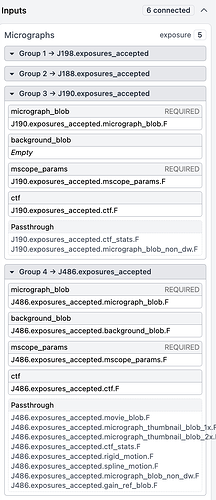
Here are the inputs from the extract job showing that one has no background_blob linked (again, these extract fine) and one does have a background_blob linked (does not extract).
This brings me to my two questions:
-
Is there a work around for this? It’s not too annoying, so for the time being I have just re-extracted all four datasets independently.
-
Will this strategy of combining data affect my overall reconstruction? Reading the other posts, it seems like the background_blob is used to estimate ice thickness across the micrograph. I assume it is therefore related to the normalization for the extracted particle. If imported aligned micros are not using this parameter, does that mean that there will be some issue down the road?
Thanks!
-Rick