Dear cryoSPARC community,
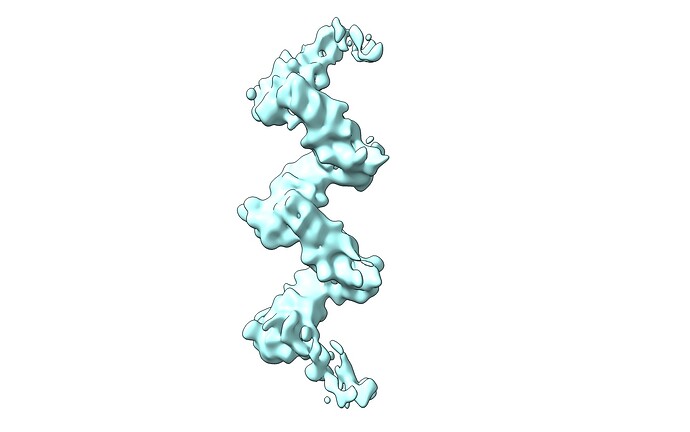
Hope you all are doing well. We are facing issues with refinement of one of our filament data sets (Protomer size=46kDa). The ab-initio reconstruction is pretty decent, and features have started to appear. The problem arises when we try to refine the structure to high resolution with helical refinement. The refined map appears to have a spiky appearance. Please refer to the parameters for helical refinement in the attachment.
Parameters for ab-initio:
Max resolution = 4A,
Initial resolution=6,
Initial mini-batch size=1000,
Final mini-batch size=3000
Parameters for helical refinement:
Helical twist=-179,
Rise=64,
Symmetry order=1,
Point group=C1,
Symmetry alignment= False,
Resolution to search helical symmetry=10,
Twist grid size=256,
Rise and grid size=10,
FSC split resolution=12,
High resolution noise substitution=True
We are using version 4.02. It would be of great help if you can suggest any change in parameters that might improve the refinement.
Thanks and Regards
1 Like
Hi @cryo-fan,
Thanks for the detailed information. Based on your parameters, the helical refinement won’t actually apply symmetry during refinement because the “Maximum symmetry order to apply during reconstruction” is set to 1. We have some information about this parameter in the Helical Refinement guide. Could you increase this parameter to at least 3, to try enforcing helical symmetry?
Best,
Michael
Dear Michael,
Thanks for your quick response. We ran the helical refinement again with “Maximum symmetry order to apply during reconstruction” set to 3, but could not see any improvement in the map or the resolution.
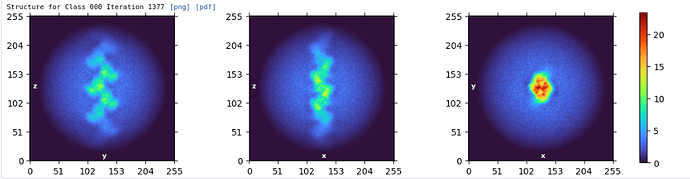
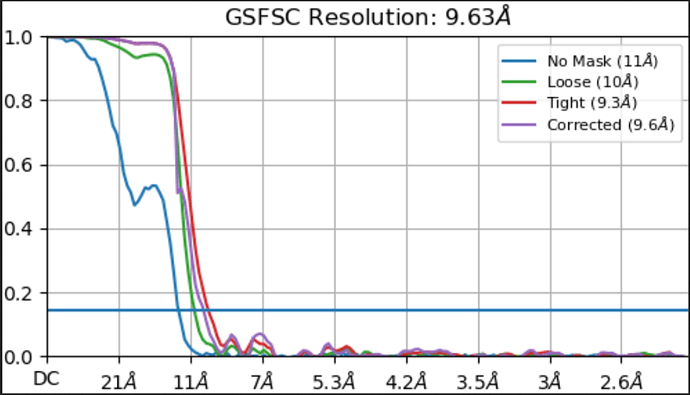
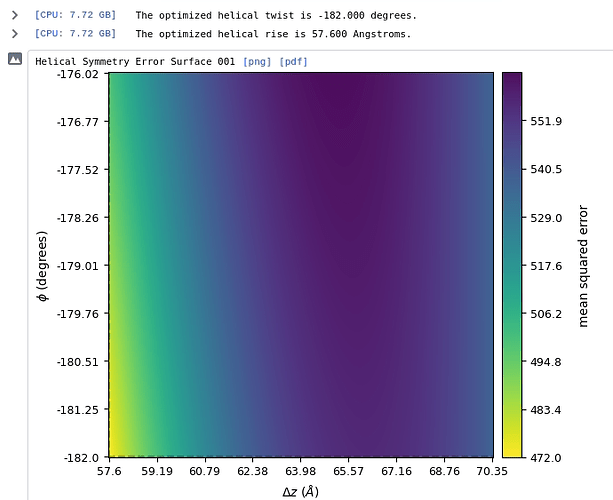
We were also wondering if the helical search parameters are correct. Please find the attached FSC plot and helical search parameters. Any suggestions with respect to the refinement and helical search parameters would be of great help.
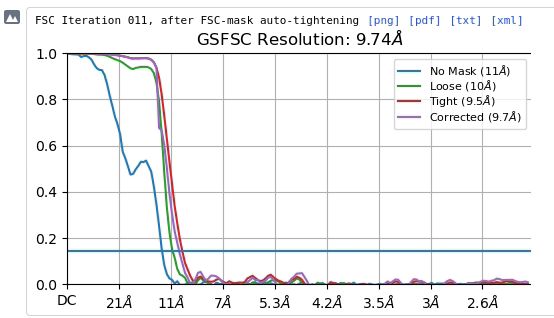
for your reference.
Thanking you
Hi @cryo-fan,
Apologies for the long delay. Based on the symmetry error plot, its likely that the helical symmetry parameters are not close enough to the correct values. Generally, if the optimization converges to edgepoints of the symmetry parameter domain, that is a warning sign that the symmetry parameters are wrong. It may be worth it to investigate whether Fourier-Bessel indexing from a high quality 2D class could help find more promising candidate symmetry parameters. (If its helpful, you could refer to external softwares for indexing, such as PyHI or HI3D.)
Best,
Michael