Hi,
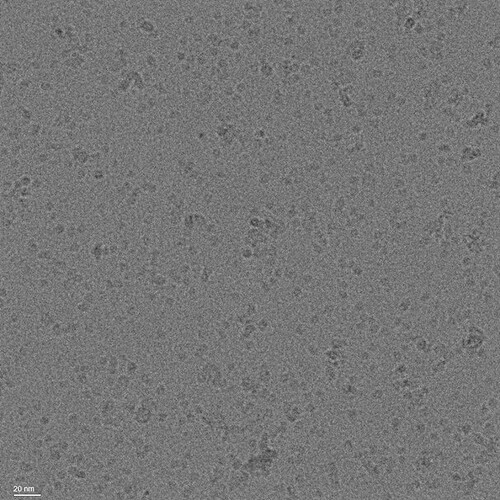
We attained micrographs with recognizeable particles. The sample is of a protein complex attained by a pull-down assay on a flag-tagged 35 kDA membrane protein solubilized in GDN. SDS page indicated that this protein is the main componenet of this complex. Here is a representative micrograph:
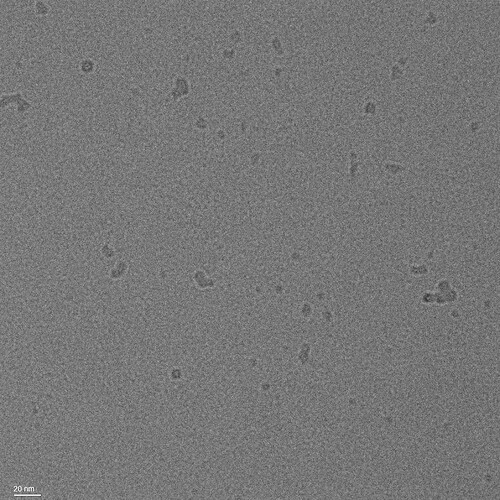
We also imaged a grid containing the buffer alone, to verify that the particles are not detergent artifacts or micelles:
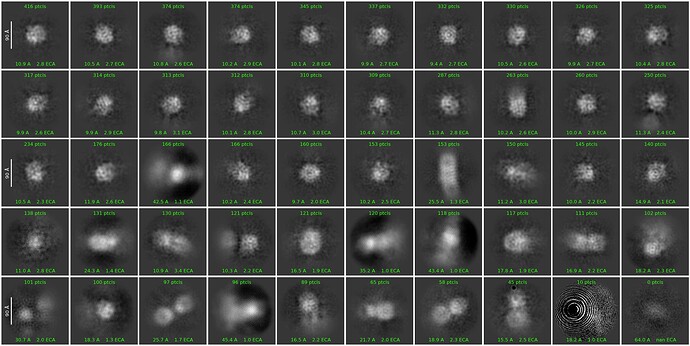
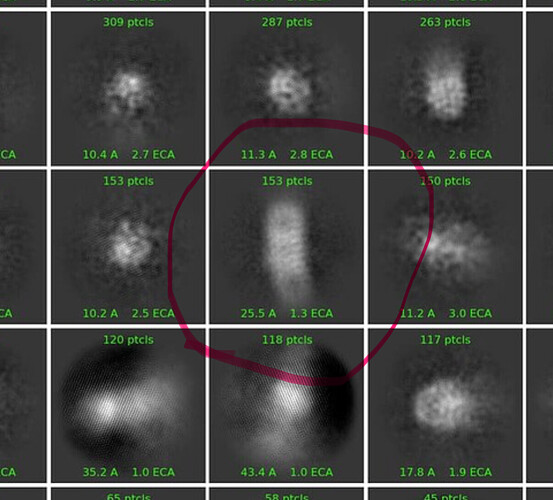
We ran 2D classification, assigning an uncertainity value of 10, picking particles from 50 to 100 angstrom. In the 25th iteration, we started to observe recognizable protein features:
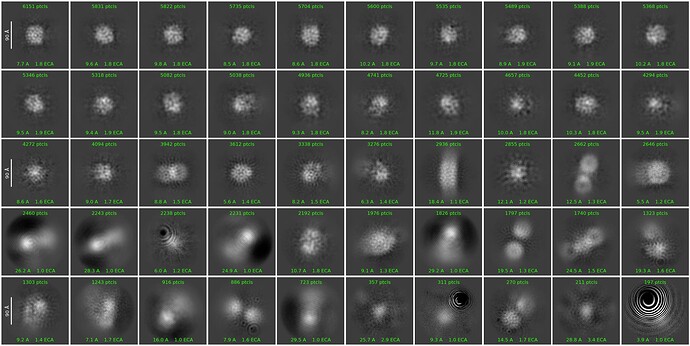
However, those were lost as iterations moved on, and particles became blurry. Here is the final (42nd) iteration:
What could have gone wrong?
Thanks alot!
Hi OrBrull,
I would try turning on solvent flattening and non-negativity to see if that helps.
These classes do not look very similar to membrane proteins that I am familiar with.
When you say you imaged buffer alone, remember that that may not be reflective of the actual micelle concentration in your sample, as GDN micelles will likely be retained even by a 100kDa spin concentrator (unless you also concentrated the buffer alone, in which case disregard this).
How much did you concentrate your sample prior to loading on the grid?
Hi,
I also concentrated the buffer alone.
I concentrated the sample after Mals-sec separation (size was estimated to be ~400kD). I concentrated the 1ml fraction to about 20ul, so about X50, in a 100k spin concentrator, to reach a concentration of about 2 mg/ml. And the buffer alone was concentrated about the same.
Perhaps this is too much?
Thanks! I will try and see if it helps
I could see this class being a side view of your protein - the others do not look like a 400kDa membrane protein to me.
What box size are you using for extraction?
Cheers
Oli
Hi,
Box size was 227A
256 pixels, 0.89A per pixel
Possibly a little small for something of this size. I would try 320px or perhaps 384px (assuming 400kDa is the total mass with detergent).
Thanks alot!
actually 400kDA is the protein mass according to MALS-SEC, total mass is about 900kDa.
And average hydrodynamic radius of the total particle was calculated to be 5nm.
So 320px should be good?
I would be extremely surprised if a 900kdA membrane protein detergent complex is actually 50 Å wide. For 900kDa, yes 320 or 384px (binned appropriately) would be a good starting point.
Looking at your mics & classes, I think you are mostly seeing micelles, with perhaps a few intact particles. You might try concentrating less, and using graphene/graphene oxide or thin carbon grids
1 Like