Hi everyone,
I solved a protein structure that has a preferred orientation issue. I don’t know if this issue can be addressed by balancing the number of particles for each view, either by increasing the particles with underrepresented views or reducing the particles with overrepresented views? If so, how can I decide whether I need to add more particles with underrepresented views instead of reducing the particles with overrepresentation views, and vice versa?
Specifically, in my case, there are fewer views in the azimuth direction between −π/2 and −π/4 and between π/4 and π/2.
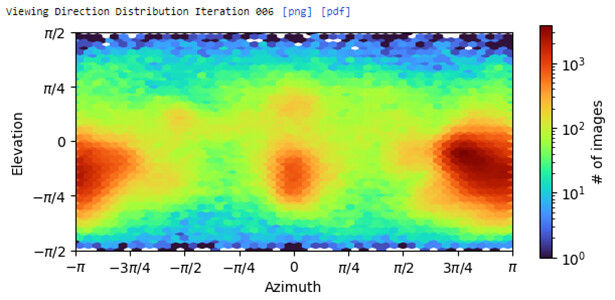
To clearly show the distribution of each view, I used the pyem program to generate the .bild file and checked the information in ChimeraX software.
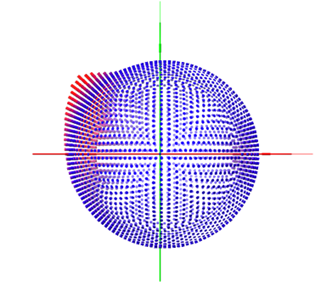
Based on the .bild file information as shown above, I am wondering if there is any existing method that can use this information to automatically balance the number of particles for each view?
Please let me know if you have any ideas and insights, any help you can provide would be highly appreciated.
Thanks,
Jianming
This seems to be a common issue and some people would say go back to sample prep.
If you think the rare views are there, do you see them in 2D classes ?
Also If you take that homo- or NU-refine job and run orientation diagnostics (https://guide.cryosparc.com/processing-data/tutorials-and-case-studies/tutorial-orientation-diagnostics) do you see the view (2D projection) with a number of 0.3 in the 3D-FSCs ? Furthermore, how does the “sphere” look in this job, is it complete or missing ?
Is there a difference in cFAR in homo vs. NU refine ?
If it is in the 2D classes you can try Rebalance 2D Classes https://guide.cryosparc.com/processing-data/all-job-types-in-cryosparc/particle-curation/job-rebalance-2d-classes-beta
I have had less luck with Rebalance Orientations https://guide.cryosparc.com/processing-data/all-job-types-in-cryosparc/particle-curation/job-rebalance-orientations but could be worth a try.
These CryoSparc jobs can help for rebalancing, but you will always have to sacrifice some particles. *you have the “rare” views there, it is not all blue.
1 Like
Rebalance orientations worked well for me recently. Play with the percentage selected, for one dataset a 92% cutoff worked best, for another, an 89% cutoff.
That preferred orientation isn’t actually too bad, it’s fairly well spread… of course the real test is the quality of the map. But I’ve seen much worse (usually from viral spike proteins…)
2 Likes

