Dear all,
I am trying to solve the structure of a protein with molecular weight of approximately 300kDa, that is predicted to be flexible and probably contains disordered regions. The particle size should be around 13 nm. Our micrographs look great, the particles are nicely visible and can be picked easily, but they fail to form reasonable 2D classes. They are fuzzy and the resolution is poor.
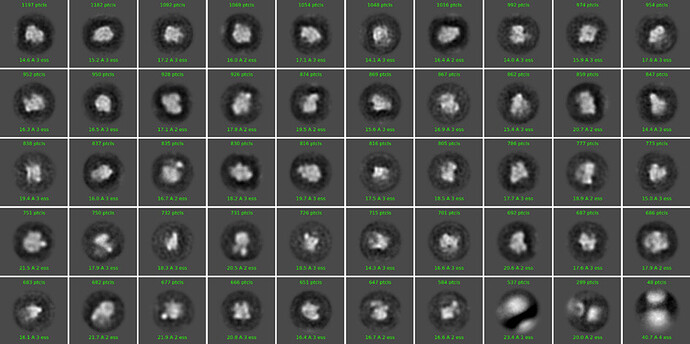
Below, you can find an example of one of our best 2D classifications. This result was obtained already after two rounds of 2D classification, from 40000 best behaving particles.
We used following parameters:
Circular mask diameter (A): 200
Circular mask diameter outer (A): 240
Force max over poses/shifts: off
Other parameters on default
Previously, I also tried to increase the batch size to 400 and number of iterations to 40, but this only led to overfitting.
Do you have any tips what could help us to improve our results?
Thank you!