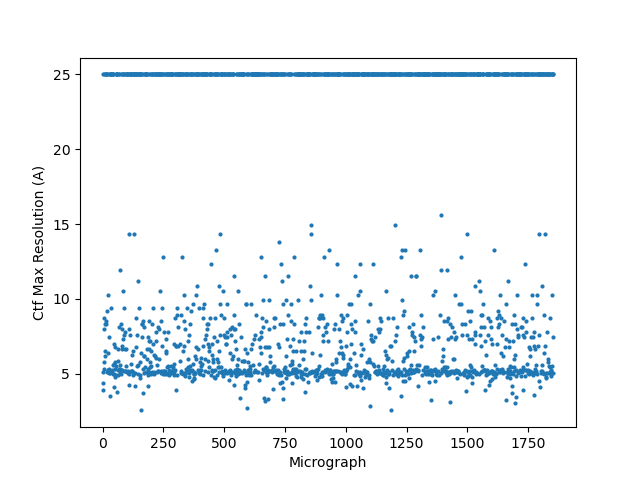
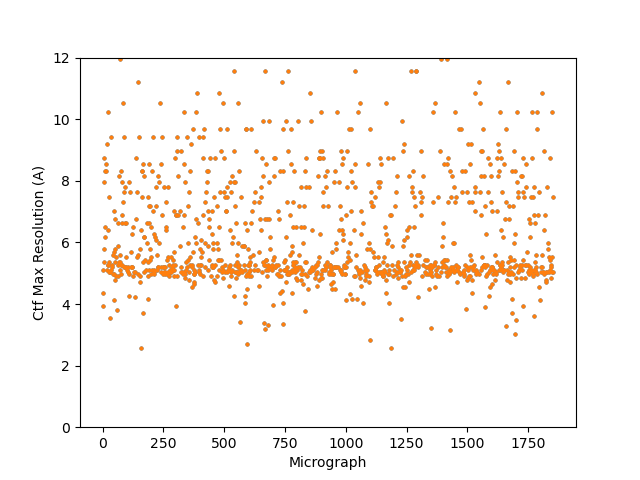
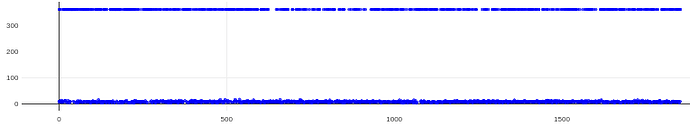
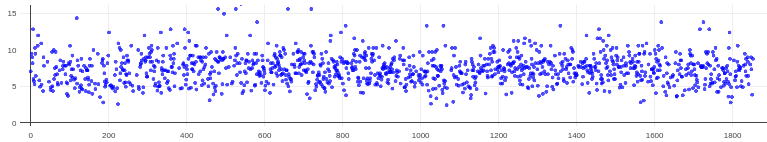
Thanks both! I tried implementing two of these suggestions with a good amount of success, results below.
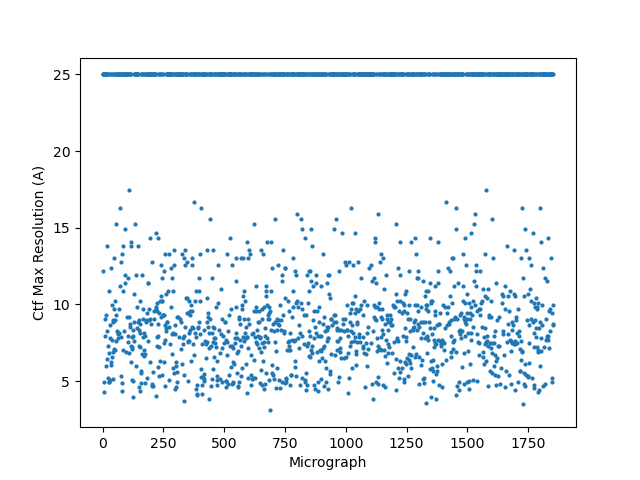
For reference, here are the original CTFs (from CSlive with default parameters) for this data set (different than those above because those above had the gold too close to the center of the micrographs so were not windowable; this was the small hole data set):
Default CTFFIND did do better:
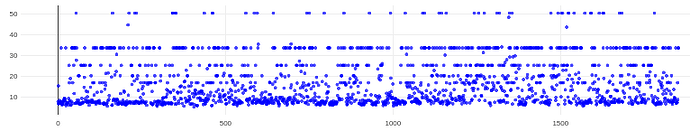
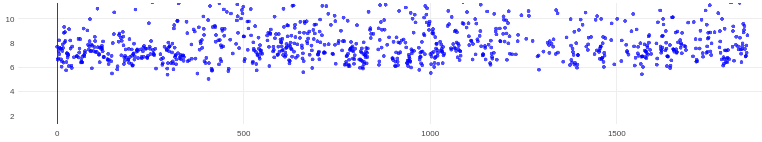
@rbs_sci (1) Relion CTF with windowing: This definitely helped!
- Estimate CTF on window size (pix): 256
- This seemed to get rid of most of the gold in the images and was still a multiple of 512; note to anyone else doing this that Relion outputs motion corrected micrographs as 512x512, not your original pixel size, so if you use the outputs of motion correction as inputs for CTFFIND, use 512 as your total length)
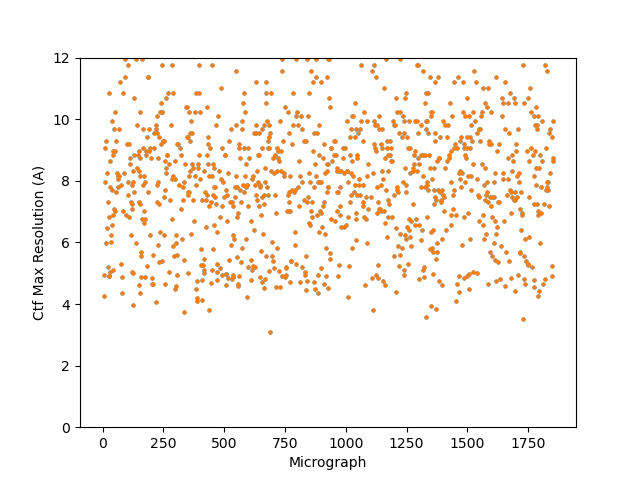
@DanielAsarnow (2) CTFFIND with altered parameters, could be done in Relion or CryoSPARC. Looked relatively similar to me as CTFFIND with default parameters. Completely makes sense on why one would want to lower minimum resolution as opposed to raise it; I think I just followed someone else’s suggestion without thinking about it first ![]()
- Lowered amplitude spectrum to 256 pixels, lowered minimum resolution to 20 A, defocus search step to 100 A.
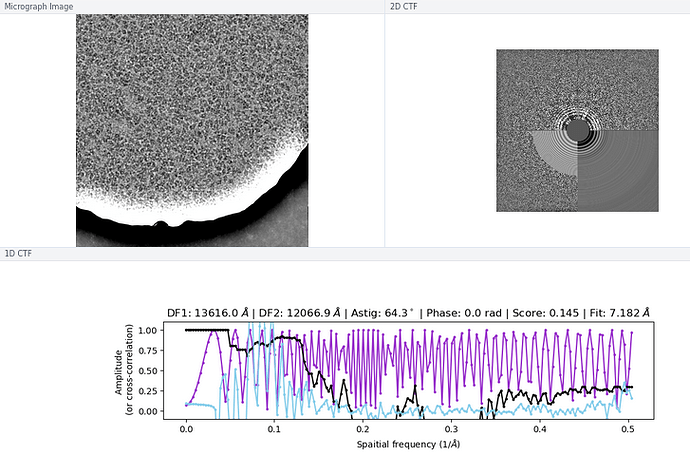
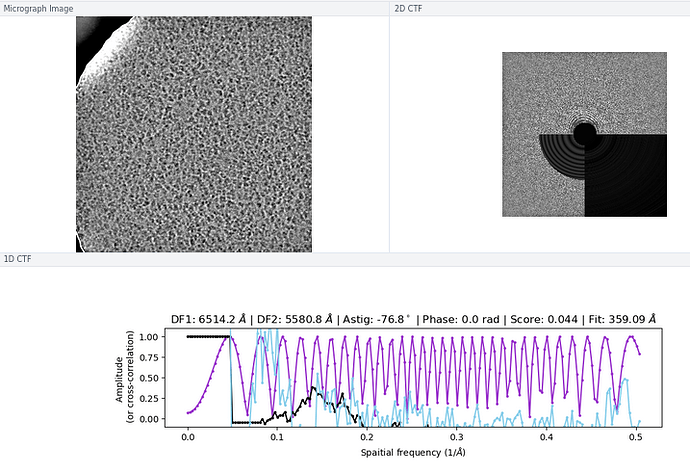
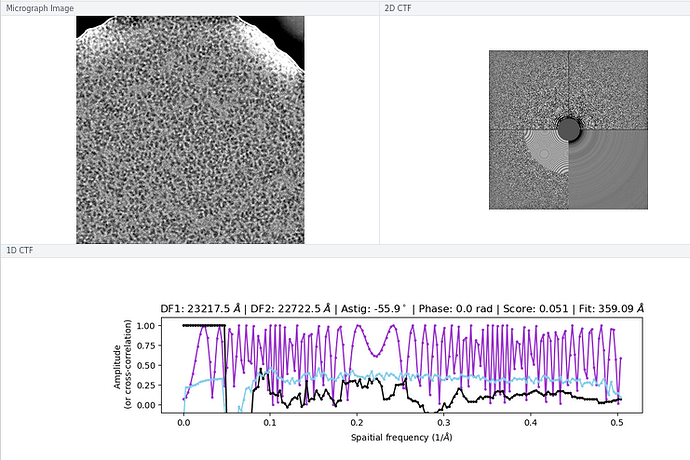
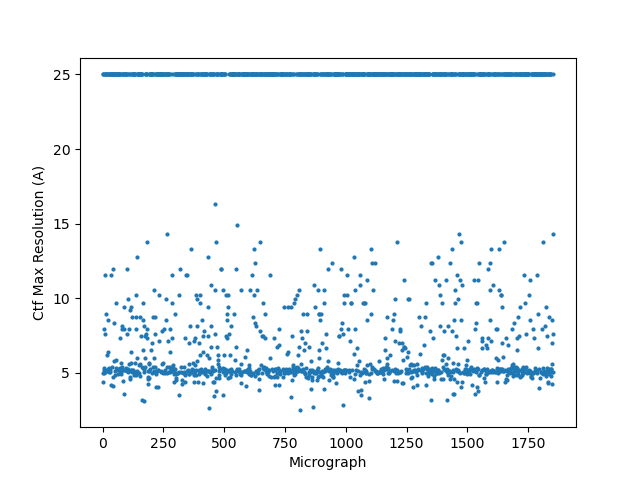
There wasn’t a super obvious reason to me why some of the micrographs still had an extremely high CTF assigned / much worse power spectra and worse rings in the CTF, although I think you are perhaps correct on the astigmatism having something to do with it (top image below is a “good” micrograph and bottom two are “bad” ones; one seems to have pretty obvious astigmatism to my eye).
(3) A combination of both (in Relion again since CS doesn’t support windowing):
- Window size 256, amplitude spectrum to 256, minimum resolution 20 A, defocus search step 100 A
- I thought this looked pretty much identical to option (1), which makes sense because if there is not much gold the other parameters probably don’t need to be changed.
- I also tried with amplitude spectrum at 128 pixels, but the results were identical to my eye
I took a look at scipion, but couldn’t find anything for masking that wasn’t just cropping borders, so if you can give a point in the right direction, I’ll look again (although as the number of programs needed increase, the willingness of my users to go through a protocol dramatically decreases).
List of feature requests for this issue:
- Bumping the request to be able to import micrographs with CTF from a star file
- Ability to window CTFs (ideally with an option to set the center of the window not in the center) and/or mask gold edges of the image