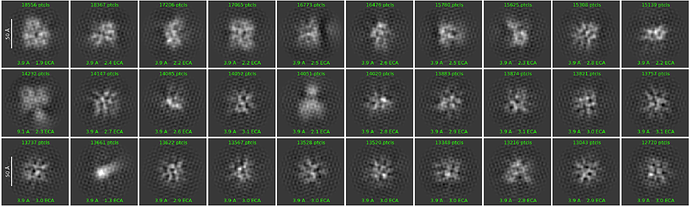
Hello, everyone.When when conducting 2D classification, it was found that the image was not clear, It’s like there’s a shadow on the grain, the molecular weight of the protein was 150KD, and DNA was added. The minimum diameter is set to 60A and the maximum diameter to 180A,the box size is 256 px,it’s fuzzy, may I ask if there is any way to improve 2D? The picture is 400,000 particles divided into 30 categories and parameter setting. Thank you very much for your help
Maybe try turning off “Force max over poses/shifts” for your 2D classification job. It might help you find good 2D classes.
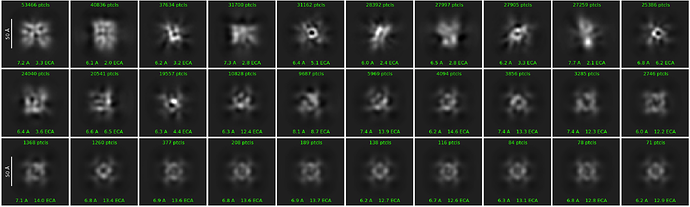
Thank you for your suggestion. According to your suggestion, I made a round of 2D again. From the image, it will be clearer, but it does not feel like protein particles.Is the particle picked inaccurate?
Your box size might be too small. You typically want your box to be 1.5-2x larger than the longest diameter of your particle (so 2*180Å=360Å) and currently with a box size of 256 px you’re at only 245 Å (256 px * 0.93Å/px). Maybe try 360 px. And a circular mask can help if you don’t think that 2D class is centering your particle correctly.
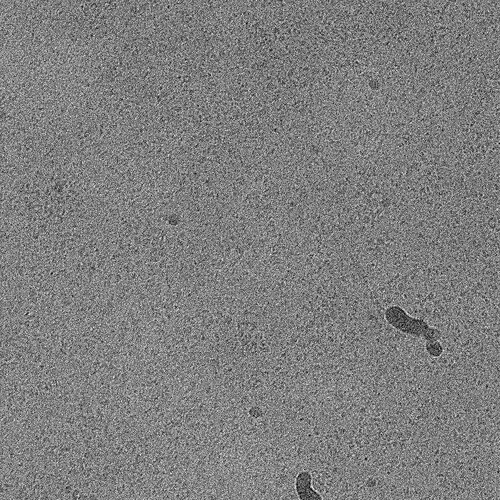
But, there’s a chance it’s not accurately picking particles. Have you run inspect particle picks after blob picking? Does it look like it’s selecting particles or noise?
Thank you for your suggestion. I’ll try it right away. I would like to ask you how to use a circular mask and how to set the corresponding parameters. Thank you for your guidance.
I just set a circular mask diameter that is slightly bigger than my particle. If you think your particle is roughly 140 Å, then do a mask of 170 Å, as an example. But it does sort of look like noise in your 2D classes right now–have you taken a look at what blob picking is giving you? Is it picking actual particles?
I would like to add to @hknox point that your 2Ds seem to have a considerable amount of noise. I’m looking at your original post and I’m wondering if your picking strategy could have some problems. From your micrographs I am unable to see any discrete particles with a recognizable and repeating shape.
I would encourage you to try two things first: 1. Inspect the particle picks of your blob picker to see if you are picking noise or actual protein; it seems like you classifying mostly noise in my opinion. 2. Do a manual picker and pick around 1K of what you consider good 2D particles and run a 2D classification (perhaps just 5 to ten classes) and see what comes up.
Ok, thank you for your suggestion. I’ll try to pick by hand.
It feels like noise now, I will try manual screening again, thank you for your tips