Hi… I realize that I put the wrong pixelsize for a dataset when I imported it. Of course, I noticed this when the CTF determination went completely crazy. Since the step of unblurring took an inordinate amount of time, I would prefer just to be able to change it after this step. Is it possible to do that, or do I have to restart the whole process?
Hi @Ruben1,
It is possible (though somewhat difficult) to change the pixel size, and also the pixel size is used to set/autotune parameters of the motion correction algorithms, so it may be worth re-processing anyway.
To change the pixel size:
- find the job directory of your motion correction job
- inside the motion correction job you should find a
passthrough_micrographs.csfile. Note the full path to this file. - run:
cryosparcm icli
# now in the cryosparc python shell
from cryosparc2_compute import dataset
d = dataset.Dataset().from_file('<path to .cs file>')
d.data['micrograph_blob/psize_A'] = <new pixel size>
d.to_file('<path to .cs file>')
This will manually override the pixel size in the output of the motion correction job, and your subsequent CTF estimation job should work correctly.
Very nice, thank you!! I reprocessed the images from the beginning, which worked fine, but this is very helpful for the future.
@apunjani, is this information still relevant for v3.2.0?
I figured out that the “cryosparc2_compute” module properly changed name to “cryosparc_compute”, but at the next step “d = dataset.from_file('<path to .cs file>')” I get this error:
AttributeError: module 'cryosparc_compute.dataset' has no attribute 'from_file'
Any idea on how to fix this?
Hey @jelka,
Sorry for the typo, it’s actually: d = dataset.Dataset().from_file('<path to .cs file>')
Thanks @stephan,
This got me to the next line, but then a new error struck.
‘mscope_params/psize_A’ does not seem exist, maybe it should be ‘movie_blob/psize_A’?
Hi @jelka,
You’re right, but there are actually a couple places where you can change the pixel size.
The pixel size result field exists in the movie_blob and the micrograph_blob result groups. The movie_blob/psize_A value will be equal to the parameter you set in the Import Movies job. The micrograph_blob/psize_A will be equal to either the parameter you set in the Import Micrographs job (if you imported micrographs directly), or if you used Motion Correction to create the micrographs, they’ll be equal to movie_blob/psize_A divided by the Fourier crop factor (the Fourier crop factor is 1 if you didn’t Fourier crop your exposures).
You can check what result fields your dataset has by printing d (e.g. print(d)). If your dataset has both, I’d suggest changing them accordingly.
Great!
Thanks for in depth explanation.
Hi, I had the same problem. I don’t know if this is useful, but my work around was to start a import micrograph job with the correct pixel size and use the the path of the dose_weighted.mrc files from the motion corrected job.
I’m trying to do this on a dataset of particles, with cryosparc-tools.
I’ve gotten as far as loading the dataset and can query the field descriptions. I see:
('blob/psize_A', '<f4'),
If I run
fe_dataset.query({
'blob/psize_A': 0.686
})
I get
Dataset([ # 879386 items, 59 fields
Which is how many particles I have. If I change that query to something my pixel size is not:
fe_dataset.query({
'blob/psize_A': 0.990
})
I get no results. So I think I’m succesfully querying the dataset.
What I’m trying to figure out is how to replace the pixel size. Naturally, I’ve settled on the dset.replace() function. The documentation states:
replace(query: Dict[str, ArrayLike], *others: Dataset, assume_disjoint=False, assume_unique=False)
There are no examples though, and I’m a bit confused about what to plug in where.
I see that:
query (dict[str, ArrayLike]) – Query description.
But what the heck is the proper syntax?
I think the closest I’ve gotten is:
fe_dataset.replace('blob/psize_A'[0.686], 0.709)
But this returns the following error:
TypeError: string indices must be integers
I assume I’m constructing the command incorrectly, but it’s not obviously to a non-programmer like myself what the syntax should be just from the function description.
I’m also wondering, how do I view the full output of the field? cryosparc-tools seems to be truncating the array:
('blob/psize_A', [0.686 0.686 0.686 ... 0.686 0.686 0.686]),
Or is that a bad idea because that’s the pixel size for every single particle in the dataset?
EDIT: I just realized that String indices must be integers means that it’s interpreting the pixel size as a string index. So let me take another crack at this.
Hi @ccgauvin94,
Thanks for the detailed post. Before attempting changing the particle’s pixel size, we wanted to ask a few questions:
- How did you determine that the pixel size was incorrect, and how did you determine the updated value?
- Are you changing the particle blob pixel size because you’d like to do additional downstream processing at the new pixel size in CryoSPARC, or would you just like to obtain half maps and FSC curves that have been calibrated to the correct pixel size?
It is true that all of the CTF parameters are linked to the particle blob pixel size. Thus, changing the blob pixel size will disturb the CTF values unless additional steps are taken.
For small changes to the pixel size, it may be possible to manually use cryoSPARC tools to update the ‘blob/psize_A’, and then to re-refine defoci via running the local CTF refinement job. If higher order aberrations were initially refined too, they will need to be re-fitted through global ctf refinement. In both of these steps, you will need to create a volume and mask that have the new updated pixel size as well, which can also be done through cryoSPARC tools.
For large changes to the pixel size (more than say, a few percent), the best approach is to start processing from scratch again unfortunately. This is because many other steps during preprocessing are calibrated based on the pixel size, and also because large changes to the pixel size are hard to “rescue” at the per-particle defocus refinement stage, once the CTF values have been fitted to it.
For information about using tools to export a new dataset with the updated pixel size, it’s worth reviewing the documentation at tools.cryosparc.com . In particular, our example on recentering particles (1. Re-center Particles — cryosparc-tools) contains useful information about how to overwrite a dataset’s field with a different value, and how to save the new dataset to CryoSPARC.
Best,
Michael
Thanks Michael,
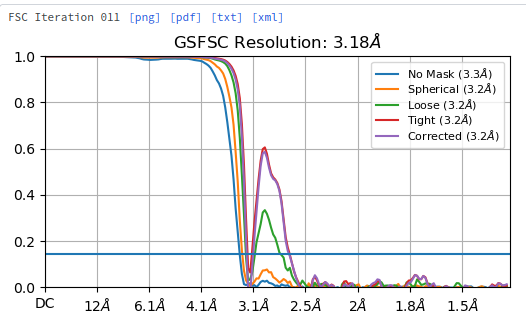
The data were collected, imported, and originally processed with 3.3% error in the pixel size. We identified the pixel size issue in a different sample by recognizing that Cs significantly refines away from the published value, and also there appears to be a potential phase inversion somewhere around 3.1 Å that leads to a complete cancellation of signal, before it then bounces right back:
For small changes to the pixel size, it may be possible to manually use cryoSPARC tools to update the ‘blob/psize_A’, and then to re-refine defoci via running the local CTF refinement job. If higher order aberrations were initially refined too, they will need to be re-fitted through global ctf refinement.
This is what I was hoping to achieve, because picking was somewhat laborious. I figured it was also a good excuse to learn more about the .cs files. My plan was to reset Cs values back to 2.7, update pixel sizes, re-scale the existing volume run local-defocus estimation, and re-refine. If it doesn’t work well, then we’ll go back to the motion correction stage.
Thanks, I’ll go through that tutorial. Really appreciate the reply!
Hi @ccgauvin94,
Did you by any chance compiled your workflow step-by-step in a document and would like to share it here?
It could be nice too many here, including me ![]()
Have a great weekend!
André
Hi Andre,
I never wound up finishing this because preliminary processing showed me that it was actually not a pixel size error. I’m still not sure what it is, but re-processing with the correct pixel size did not solve the issue.
It looks very much like what we see in cases of severe optical optical aberrations, e.g extreme beamtilt - have you tried Global CTF either with one group, or with image shift groups?
Yes - weirdly the issue is only for this sample. Multiple data collections of this sample all exhibit it, while other samples don’t. And once I get past ~2.5 Å resolution, the aberration goes away.
That’s a very interesting phenomenon! Whenever you reach more conclusions, keep us posted.
I’ve been using magCalEM from Chris Russo’s group to obtain the correct pixel size, and just reprocessing by re-import, patch ctf, re-associate particle to mics, re-refine.
@ccgauvin94 how is that particle shaped?
It’s a miniferritin with tetrahedral symmetry. It has a hollow core that is about 4 nm. The problem seems like it might be more pronounced in datasets that have iron bound in the core, but I haven’t reached any firm conclusions yet.
Hi Daniel (@DanielAsarnow),
Thank you so much for sharing your experience!
I have been wanting to try magCalEM for some time, but I haven’t yet had the chance to do so. If I understand correctly, magCalEM makes use of gold foils to estimate the pixel size, rather than other methods that iterativelly rescale the pixel/voxel size of the cryo-EM map to find the best cross-correlation to a reference map.
Using magCalEM one can estimate the pixel size before data processing, but the workflow you described made me think that you are revisiting previous datasets.
Would you be happy sharing your experience and a perspective of how much the quality of the final 3D volume of some of those datasets may have improved?
Happy holidays to everyone! =)
André