Hi,
This is more of an observation than anything - more of a comment than a question as it were  - but I figured it might be useful to share with the community.
- but I figured it might be useful to share with the community.
Basically, I’ve noticed that in datasets with significant heterogeneity, if I subset particles from the consensus refinement based on the per particle scale, different scale factor bins have different conformations.
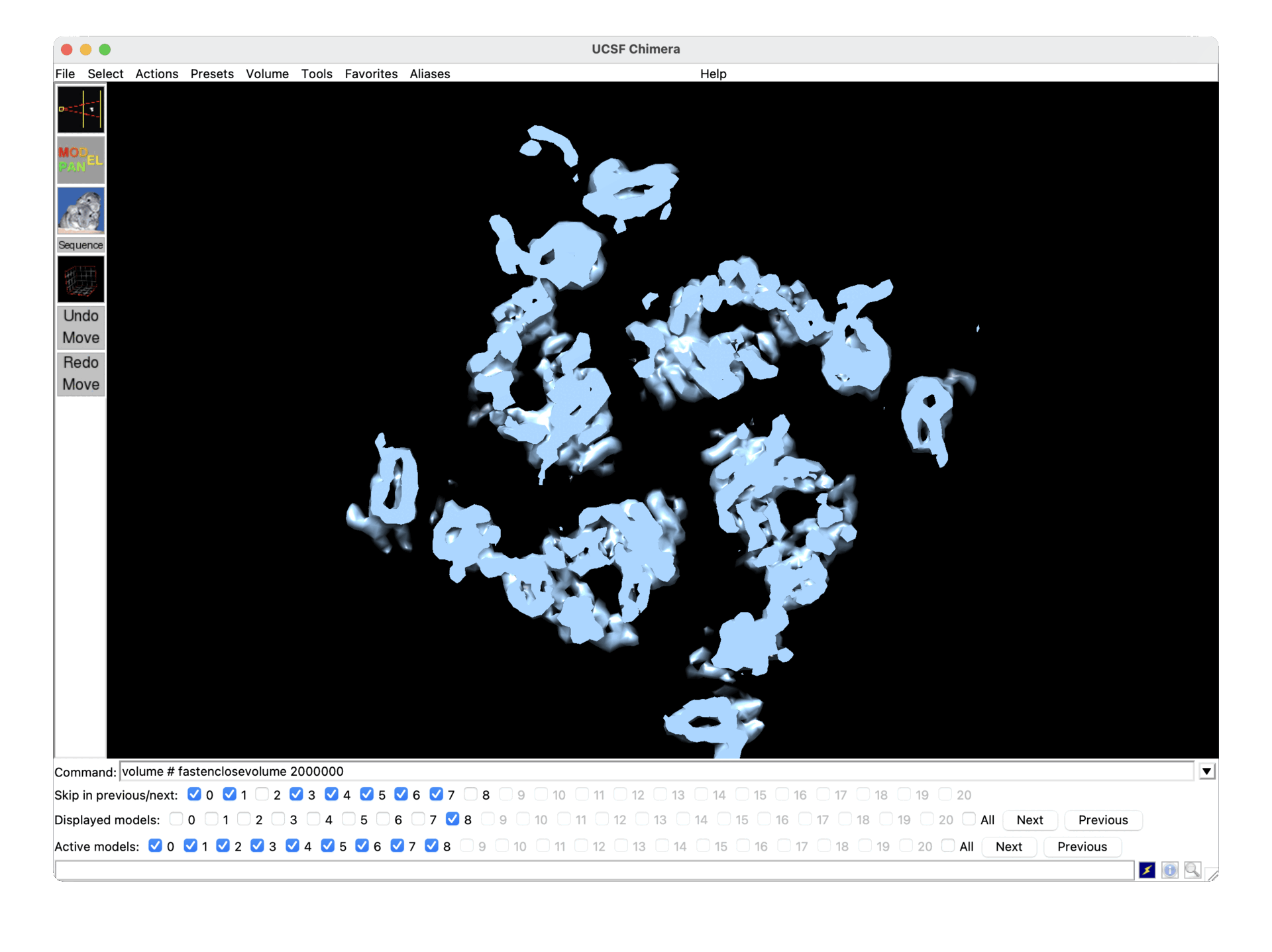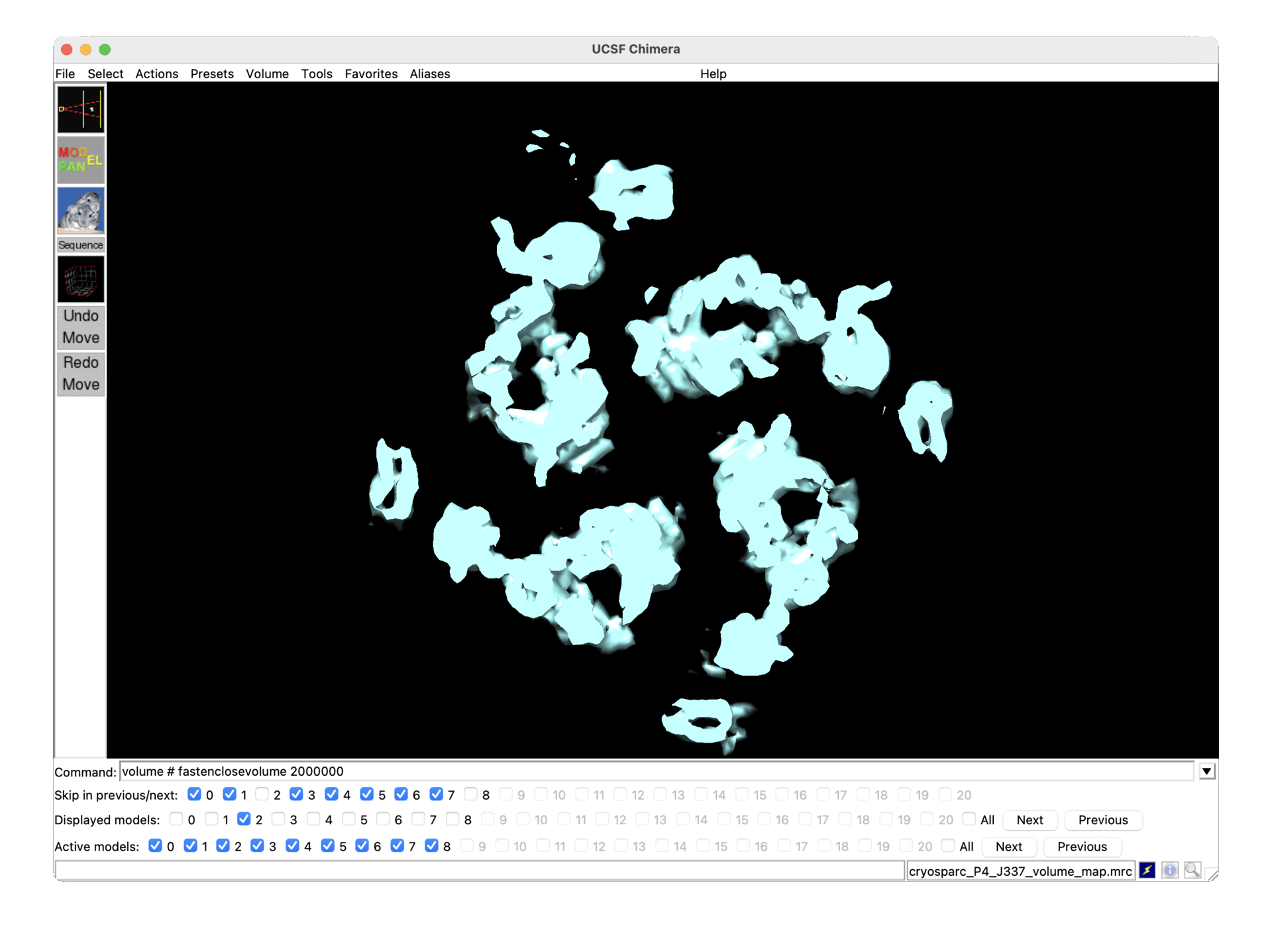
This is mentioned briefly in the 3D classification docs in the context of compositional heterogeneity, but it also pertains to conformational heterogeneity, as you can see below (one state in the gif is scale ~0.7, the other ~1.2, both refined together and then reconstructed separately after Subset by Statistic).
Presumably in cases like this, it would be best to turn the use of scale factors off in 3D classification and 3D-VA?
Cheers
Oli
3 Likes
Hey @olibclarke – this is super interesting! Have you (or anyone else reading this) seen this scale-based conformational classification work in other data? We’ve been playing with subsetting by scale internally, but largely for binary accept/reject schemes.
Presumably in cases like this, it would be best to turn the use of scale factors off in 3D classification and 3D-VA?
We would be hesitant to recommend this generally because per-particle scale can capture quite a bit of variation in signal magnitude that is not linked to real heterogeneity (e.g., ice thickness, different per-micrograph normalization, etc.). Thus turning off per-particle scale minimization might result in many modes/classes focussing on this irrelevant dimming/brightening. However, worth a shot! Please report back if you see something notable.
One additional note: naming this scalar multiple a ‘scale’ might obscure some of its use in cases like these. Another way one might think about this number is as a ‘scaled correlation’. That is, the per-particle scale is exactly equal to the correlation between slice and image, multiplied by the ratio of their magnitudes. It can be high / low either because the two magnitudes differ, or because the correlation is high / low.
1 Like
Hi @vperetroukhin,
I haven’t tried with other data, but thinking about this:
One additional note: naming this scalar multiple a ‘scale’ might obscure some of its use in cases like these. Another way one might think about this number is as a ‘scaled correlation’. That is, the per-particle scale is exactly equal to the correlation between slice and image, multiplied by the ratio of their magnitudes. It can be high / low either because the two magnitudes differ, or because the correlation is high / low.
In a case like this, my guess is that because this is a huge molecule, there is a tonne of signal, and so effects of correlation (due to heterogeneity) are larger?
Certainly in this case, turning off scale factors doesn’t seem to have any negative effects on downstream 3D-VA or Class3D that I can tell.
![]() - but I figured it might be useful to share with the community.
- but I figured it might be useful to share with the community.