Hello, we are processing eer micrographs with pixel size 0.58 binned twice on cryosparc using patch motion and 2600 images took almost 6 hours to complete. Is that normal since relion can do the motion correction of 1000 in less than 1 hour? Are we running it wrong. What is a normal speed?
EER upsampling=2 at import perhaps? This does slow things down quite a bit relative to 4K-rendering, which RELION defaults to.
Cheers,
Yang
Depends a lot on where the micrographs are, although for high mag EER data file sizes should be small enough not to be a problem.
My guess is, as @leetleyang also hypothesised, that you left EER sampling at 2. This will output 8K movies, which isn’t the issue… the bigger issue will be that CryoSPARC struggles with CTF estimation at very small pixel sizes, so chances are Patch CTF Estimation will fail.
It’s interesting, because I usually find RELION motion correction (using “RELIONCor”) is significantly slower than Patch Motion Correction of EER in CryoSPARC…
I am not upsampling. We did patch motion on 9000 images in 20 hours, pixel size 0.58, 45 e/A dose.
Is that normal when I get 1000 movies/hour in Relion?
I see.
Can you confirm the number of GPUs you’re parallelising Patch Motion Correction across, and in comparison, the MPI/thread values, nodes, and cores-per-node in your RELION scenario?
Cheers,
Yang
I am using 1 GPU for the patch motion and for relion I am using 32 MPI/20 threads, no GPUs.
would using more CPUs make the jobs in cryosparc faster?
Thank you. In that case, the throughput you’re seeing is quite normal, on both fronts.
Your RELION parameters instruct RelionCor to process 32 movies in parallel (each in 20-frame batches). What CPU-processing loses out in compute, it makes up for in multithreadedness and parallelisation. I assume the MPIs are spread across several nodes in your cluster, which alleviates network I/O bottlenecks.
On the cryoSPARC front, 9000 EER moviestacks over 20 hours works out to 7.5 stacks/min, which feels normal to me given the 1 GPU-assignment. The good news is that this will scale somewhat linearly with the number of GPUs you assign to the task–within reason. Try increasing Number of GPUs to parallelize to 2 or 4 (depending on your workstation/node configuration) and see if that helps.
Cheers,
Yang
We have limited number of tokens and cannot really afford to use 4 GPUs. Is there any way to accelerate the process by increasing the threads in the cryosparc jobs and not making is so realiant on GPUs?
Unfortunately not. All compute-heavy tasks in cryoSPARC are, by design, GPU-accelerated.
You can limit downtime by preprocessing movies in cryoSPARC Live as they’re being generated/transferred. A modern GPU would be able to keep pace with typical collection throughputs.
Another option is to perform motion correction in RELION and import the averaged micrographs into cryoSPARC for downstream processing. I believe the community has generally found Patch Motion Correction to do a better job, for whatever reason, but RelionCor is perfectly workable. However, this workflow will preclude Reference-Based Motion Correction. As Sanofi seems to have access to a lot CPU resources, Bayesian polishing would be a viable alternative.
Cheers,
Yang
Confirming the post from @leetleyang, CryoSPARC’s Patch motion correction is GPU accelerated and does not have a CPU-only option. Parallelism over multiple GPUs generally provides a near-linear performance increase unless there are other bottlenecks such as the filesystem. Performance will depend on a number of factors:
- For EER data,
EER Upsampling Factorparameter (default 2x) causes the data to be processed in 2x super resolution. We set this default in order to be conservative and ensure that the available signal in the EER files can be made use of downstream. However, in some cases a 2x upsampling factor may not be required and can be set to 1x instead. This is especially the case if micrographs will be downsampled viaOutput F-crop factoranyway. Changing to 1x reduces the number of pixels processed during motion correct by a factor of 4. - Filesystem performance can have a large impact. You can see the relative times of loading vs. processing data in the output of Patch motion jobs, e.g.:
-- 0.0: processing 2 of 20: J1/imported/14sep05c_00024sq_00004hl_00002es.frames.tif
loading /bulk6/data/cryosparcdev_projects/P103/J1/imported/14sep05c_00024sq_00004hl_00002es.frames.tif
Loading raw movie data from J1/imported/14sep05c_00024sq_00004hl_00002es.frames.tif ...
Done in 2.26s
Loading gain data from J1/imported/norm-amibox05-0.mrc ...
Done in 0.00s
Processing ...
Done in 6.99s
Completed rigid and patch motion with (Z:5,Y:8,X:8) knots
Writing non-dose-weighted result to J2/motioncorrected/14sep05c_00024sq_00004hl_00002es.frames_patch_aligned.mrc ...
Done in 0.39s
Writing 120x120 micrograph thumbnail to J2/thumbnails/14sep05c_00024sq_00004hl_00002es.frames_thumb_@1x.png ...
Done in 0.31s
Writing 240x240 micrograph thumbnail to J2/thumbnails/14sep05c_00024sq_00004hl_00002es.frames_thumb_@2x.png ...
Done in 0.00s
Writing dose-weighted result to J2/motioncorrected/14sep05c_00024sq_00004hl_00002es.frames_patch_aligned_doseweighted.mrc ...
Done in 0.38s
Writing background estimate to J2/motioncorrected/14sep05c_00024sq_00004hl_00002es.frames_background.mrc ...
Done in 0.01s
Writing motion estimates...
Done in 0.00s
- Different GPU models will have different performance. Typically GPU memory bandwidth has the most significant impact on performance
Hope this helps!
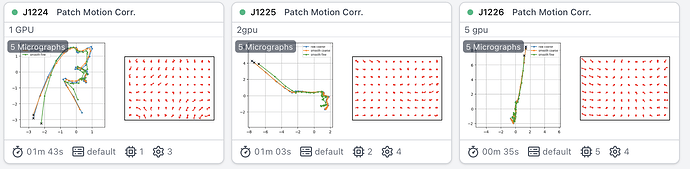
I have a similar problem of speed.
I tested running patch motion with 1 2 and 4 gpus with puzzling results:
1.5min for 5 movies with 1 gpu, more than 5 min with 2gpus, experiment canceled with 4 gpus as it was even longer.
I have A6000 GPUs on a very solid system with a ton or RAM, but something seem to be jamming it.
I noticed a similar behavior with Patch CTF, and also with NU-refine.
If I run only one NU-refine job with ~20k particles, it takes ~2min to run.
As soon as I run a second NU-refine job at the same time on a different GPU, the run time jumps to 10 min. More than 2 jobs, hours. I am running cryosparc v4.6.0
@adesgeorges Please can you post the output of
- this command on the CryoSPARC master computer
cryosparcm cli "get_scheduler_targets()" - this command on the GPU server
cat /sys/kernel/mm/transparent_hugepage/enabled free -h
cryosparcm cli "get_scheduler_targets()"
[{'cache_path': '/ssd/cryosparc_cache/', 'cache_quota_mb': None, 'cache_reserve_mb': 10000, 'desc': None, 'gpus': [{'id': 0, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 1, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 2, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 3, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 4, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 5, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 6, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}, {'id': 7, 'mem': 51041271808, 'name': 'NVIDIA RTX A6000'}], 'hostname': 'headnode', 'lane': 'default', 'monitor_port': None, 'name': 'headnode', 'resource_fixed': {'SSD': True}, 'resource_slots': {'CPU': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103], 'GPU': [0, 1, 2, 3, 4, 5, 6, 7], 'RAM': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63]}, 'ssh_str': 'cryosparc_user@headnode', 'title': 'Worker node headnode', 'type': 'node', 'worker_bin_path': '/spshared/apps/cryosparc4/cryosparc_worker/bin/cryosparcw'},
{'cache_path': '/ssd/cryosparc_cache', 'cache_quota_mb': None, 'cache_reserve_mb': 10000, 'custom_var_names': [], 'custom_vars': {}, 'desc': None, 'hostname': 'headnode_slurm', 'lane': 'headnode_slurm', 'name': 'headnode_slurm', 'qdel_cmd_tpl': 'scancel {{ cluster_job_id }}', 'qinfo_cmd_tpl': 'sinfo', 'qstat_cmd_tpl': 'squeue -j {{ cluster_job_id }}', 'qstat_code_cmd_tpl': None, 'qsub_cmd_tpl': 'sbatch {{ script_path_abs }}', 'script_tpl': '#!/bin/bash\n#SBATCH --job-name=cryosparc_{{ project_uid }}_{{ job_uid }}\n#SBATCH --partition=A6000\n#SBATCH --output={{ job_log_path_abs }}\n#SBATCH --error={{ job_log_path_abs }}\n{%- if num_gpu == 0 %}\n#SBATCH --ntasks={{ num_cpu }}\n#SBATCH --cpus-per-task=1\n#SBATCH --threads-per-core=1\n{%- else %}\n#SBATCH --nodes=1\n#SBATCH --ntasks-per-node={{ num_cpu }}\n#SBATCH --cpus-per-task=1\n#SBATCH --threads-per-core=1\n#SBATCH --gres=gpu:{{ num_gpu }}\n#SBATCH --gres-flags=enforce-binding\n{%- endif %}\n\navailable_devs=""\nfor devidx in $(seq 0 7);\ndo\n if [[ -z $(nvidia-smi -i $devidx --query-compute-apps=pid --format=csv,noheader) ]] ; then\n if [[ -z "$available_devs" ]] ; then\n available_devs=$devidx\n else\n available_devs=$available_devs,$devidx\n fi\n fi\ndone\nexport CUDA_VISIBLE_DEVICES=$available_devs\n\n{{ run_cmd }}\n', 'send_cmd_tpl': '{{ command }}', 'title': 'headnode_slurm', 'tpl_vars': ['num_gpu', 'num_cpu', 'project_uid', 'job_log_path_abs', 'cluster_job_id', 'command', 'job_uid', 'run_cmd'], 'type': 'cluster', 'worker_bin_path': '/spshared/apps/cryosparc4/cryosparc_worker/bin/cryosparcw'}]
cat /sys/kernel/mm/transparent_hugepage/enabled
[always] madvise never
This setting is problematic. To confirm its impact, you can test if multi-GPU or multi-job performance improves if you disable transparent_hugepage with the commands
sudo sh -c "echo never > /sys/kernel/mm/transparent_hugepage/enabled"
This command would disable transparent_hugepage temporarily. See here for how to change the setting permanently.
CryoSPARC v4.6.2 includes a workaround that is effective when
cat /sys/kernel/mm/transparent_hugepage/enabled
shows
always [madvise] never.
May I ask: in what situations having this parameter set like this might be detrimental? Why would it be enabled as default if it is so damaging to performance?
Because it’s not always damaging to performance, and can for some things significantly improve performance and memory handling. When a CPU accesses memory, it examines the Translation Lookaside Buffer (TLB) to find where it needs to look (which is stored on the CPU) and a “miss” in the caching can have significant impact as they it needs to look in main system RAM (which is much slower than on-die CPU cache). Reducing the number of misses improves performance.
I don’t know how much of a rabbit hole you want to go down, but some things (like the Redis database, and CryoSPARC itself) have severe performance regressions with THP enabled. In fact, databases in general seem to see significant loss of performance, which if I understand it correctly is likely due to the fact that databases rarely read memory contiguously, but are accessing in a much more stochastic fashion. I went down this rabbit hole myself a few months ago when it became obvious that THP was a pain point for CryoSPARC - I wasn’t sure whether it was database related or whether cryo-EM data processing in general was impacted, but I haven’t seen RELION act as sensitively as CryoSPARC to THP being enabled or not.
This site has a reasonable rundown if you just want a one page read to sate curiosity. ![]()
hey @adesgeorges,
This improvement is just because of updating to 4.6.2 or did you also disable the transparent_hugepage ?
Thanks
I did not update, only disabled transparent_hugepage.