Hi
Thanks for your response ![]() The details below.
The details below.
the text (as text
) of the traceback
The first error that is displayed (for second processed micrograph):
Error occurred while processing J8/imported/002081751992695112843_20220713_10096_RawImages_resized.mrc
Traceback (most recent call last):
File “/home/schubert/cryosparc/cryosparc_worker/cryosparc_compute/jobs/pipeline.py”, line 60, in exec
return self.process(item)
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 112, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 118, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 119, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/patchctf.py”, line 70, in cryosparc_compute.jobs.ctf_estimation.patchctf.patchctf_v217
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/patchctf.py”, line 116, in cryosparc_compute.jobs.ctf_estimation.patchctf.patchctf_v217
File “/home/schubert/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/pycuda/gpuarray.py”, line 550, in fill
func.prepared_async_call(self._grid, self._block, stream,
File “/home/schubert/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/pycuda/driver.py”, line 565, in function_prepared_async_call
arg_buf = pack(func.arg_format, *args)
struct.error: required argument is not an integer
Marking J8/imported/002081751992695112843_20220713_10096_RawImages_resized.mrc as incomplete and continuing…
The second error that occurs (for micrograph 3 onward):
Error occurred while processing J8/imported/000561236593900554028_20220713_10097_RawImages_resized.mrc
Traceback (most recent call last):
File “/home/schubert/cryosparc/cryosparc_worker/cryosparc_compute/jobs/pipeline.py”, line 60, in exec
return self.process(item)
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 112, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 118, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py”, line 119, in cryosparc_compute.jobs.ctf_estimation.run.run.ctfworker.process
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/patchctf.py”, line 70, in cryosparc_compute.jobs.ctf_estimation.patchctf.patchctf_v217
File “cryosparc_master/cryosparc_compute/jobs/ctf_estimation/patchctf.py”, line 111, in cryosparc_compute.jobs.ctf_estimation.patchctf.patchctf_v217
File “cryosparc_master/cryosparc_compute/engine/cuda_core.py”, line 361, in cryosparc_compute.engine.cuda_core.EngineBaseThread.ensure_allocated
File “/home/schubert/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/pycuda/gpuarray.py”, line 782, in ravel
return self.reshape(self.size)
File “/home/schubert/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.8/site-packages/pycuda/gpuarray.py”, line 778, in reshape
gpudata=int(self.gpudata),
TypeError: int() argument must be a string, a bytes-like object or a number, not ‘NoneType’
Marking J8/imported/000561236593900554028_20220713_10097_RawImages_resized.mrc as incomplete and continuing…
- your CryoSPARC version
Cryosparc V4.2.1
- the job type of J8
Patch CTF Estimation
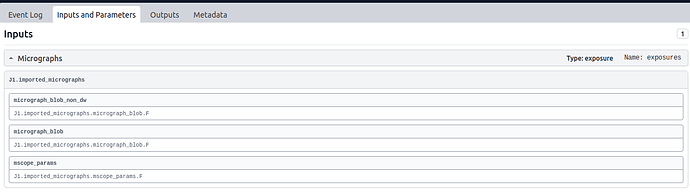
- a screenshot of the expanded Inputs under Inputs and Parameters
- any non-default parameters
All parameters set to default
- a description of the J8 inputs: provenance, dimensions
We are using micrographs taken using a DE-20 direct electron detector which has dimensions:
5090 * 3770.
Motion correction has already been performed outside of cryosparc so these are alligned .mrc micrographs. I thought the rectangular aspect ratio might be the cause so i have tried this with padded edges to achieve dimensions 5090 * 5090, however the error is the same.
I also want to note here that I migrated these micrographs to a different instance i have access to and the error repeats so it seems it’s some issue with my data.
Thanks so much for your time,
James.