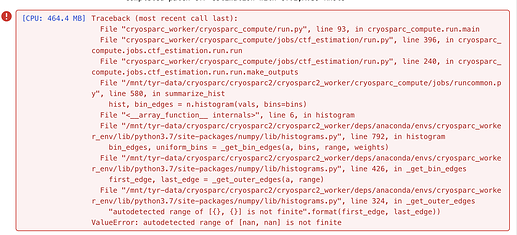
I’m getting this error for certain micrographs, I’m not 100% sure but I believe it’s just ones that have very, very low counts due to a misaligned energy filter. The Patch CTF job fails so I can’t proceed to Curate Exposures in order to discard them. (These micrographs would be useless themselves, but the job is interrupted part way through).
Thank you for reporting this issue. We have taken note of it and are considering a fix.
I don’t have a great recommendation for a temporary workaround. Have you checked already if a meaningful, preliminary curation is possible based on the motion correction results?
Motion-based curation almost definitely would have worked, but I ran Motioncor2 outside cryoSPARC this time. My workaround was to run the incomplete micrographs in CTFFIND4, and then curate those, throw away the bad micrographs (they’re almost blank, CTF fits >> 20 Å), and finally run the good ones through Patch CTF again.
Had a similar issue to @DanielAsarnow. Imported .eer, upsampled = 2 (8k), then path motion ran fine with all micrographs.
Then Patch CTF failed after ~59sec. I ran Check for Corrupt Micrographs, none detected - but Patch CTF still failed.
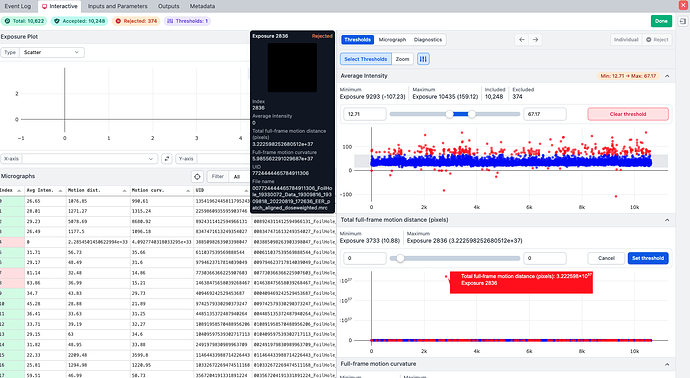
I took the micrographs from Patch Motion into a curation job, it turned out just one really bad one out of ~10,600. Once this was removed, the accepted micrographs ran fine through Patch CTF.
In the end, just one bad apple killed Patch CTF.
@Mark-A-Nakasone What version of CryoSPARC did you use? Did the Patch CTF job fail entirely, or was just the “bad” micrograph marked as failed, and all other micrographs were successfully processed?
Hi @wtempel, in this case it was on v4.6.1. Import and Patch motion were fine. Patch CTF failed to where it completely output nothing - because the “bad” micrograph was one of the first processed. It would be possible to mark the failed job as complete and take the partial micrographs.
@Mark-A-Nakasone, was your error message identical to the one posted by Daniel? “autodetected range of [nan, nan] is not finite”?
If you could paste the error message that you saw, that would be very helpful.
Hi @hsnyder, I deleted the job already.
To replicate the error, I just went back to the curated exposures after Patch Motion.
In this case I ran the rejected exposures through Patch CTF and the error is about as you say.
I think it is just the bad micrographs, maybe the transfer corrupted them.
[2025-02-28 1:40:12.47]
[CPU: 288.3 MB Avail: 180.93 GB]
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 129, in cryosparc_master.cryosparc_compute.run.main
File "cryosparc_master/cryosparc_compute/jobs/ctf_estimation/run.py", line 400, in cryosparc_master.cryosparc_compute.jobs.ctf_estimation.run.run
File "/home/cryosparcuser/cryosparcV3/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/matplotlib/pyplot.py", line 1998, in ylim
ret = ax.set_ylim(*args, **kwargs)
File "/home/cryosparcuser/cryosparcV3/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/matplotlib/axes/_base.py", line 3898, in set_ylim
return self.yaxis._set_lim(bottom, top, emit=emit, auto=auto)
File "/home/cryosparcuser/cryosparcV3/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/matplotlib/axis.py", line 1210, in _set_lim
v0 = self.axes._validate_converted_limits(v0, self.convert_units)
File "/home/cryosparcuser/cryosparcV3/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.10/site-packages/matplotlib/axes/_base.py", line 3585, in _validate_converted_limits
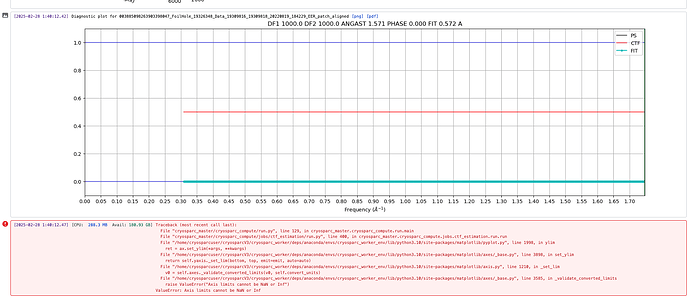
raise ValueError("Axis limits cannot be NaN or Inf")
ValueError: Axis limits cannot be NaN or Inf
Hi @Mark-A-Nakasone,
Thanks for sharing that. This is a known issue and a fix will be included in the next version of CryoSPARC. in the meantime, rejecting the exposure and continuing with the rest of the dataset is the best workaround.
– Harris