Hi all,
I am new to cryo-em and cryosparc. I am working on my master thesis with a small protein complex (about 100 kDa). The dataset was collected with phase plates and I am having some troubles with the CTF estimation.
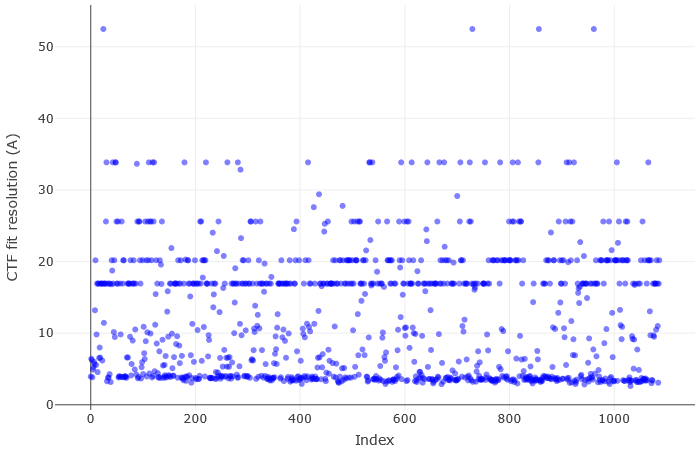
If I run patch CTF, I get some distinct CTF fit resolution “streaks”. See the plot below:
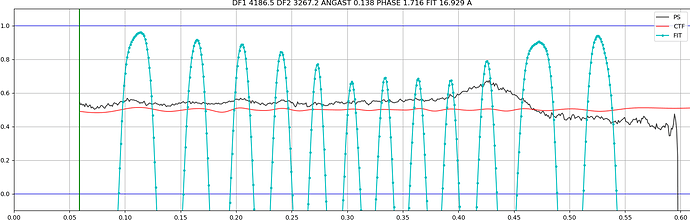
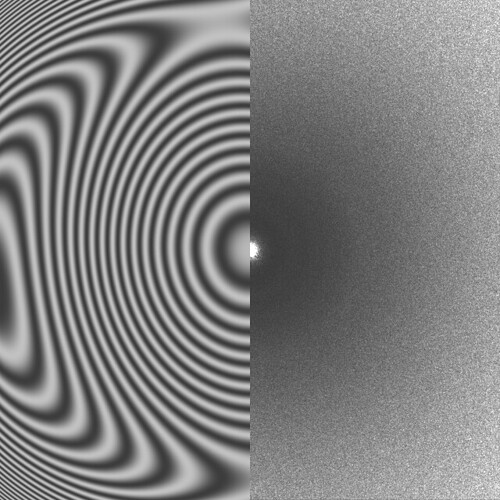
Some of them have very faint Thon rings or even not visible, which explains why the algorithm fails. However, there are some cases in which the fit is “shifted”:
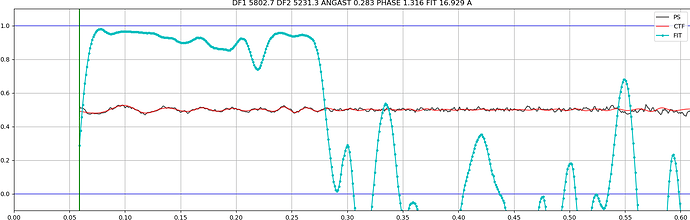
In other cases the observed CTF is similar to the fit but not at lower resolution, which makes it fail when setting the CTF fit resolution:
I tried tweaking the resolution limits when estimating the CTFs but it did not help.
Do you have any suggestions on how I could improve this? or is the signal too small?
Thank you!