Thank you wtempel!
-
additional lines preceding the error message in the event log
[CPU: 584.7 MB] – 0.0: processing 8 of 14646: J20/motioncorrected/000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned.mrc loading /mnt/nfs/user_data/Sa_Chen/cryosparc_projects/SC25_1/P27/J20/motioncorrected/000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned.mrc Loading raw mic data from J20/motioncorrected/000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned.mrc … Done in 0.03s Processing … Done in 2.34s Completed patch CTF estimation with (Y:2,X:2) knots
1D search for 007658629387853597550_FoilHole_20039935_Data_18963670_18963672_20230320_065207_EER_patch_aligned [png] [pdf]

1D search result for 007658629387853597550_FoilHole_20039935_Data_18963670_18963672_20230320_065207_EER_patch_aligned [png] [pdf]

2D patch result for 007658629387853597550_FoilHole_20039935_Data_18963670_18963672_20230320_065207_EER_patch_aligned [png] [pdf]

Diagnostic plot for 007658629387853597550_FoilHole_20039935_Data_18963670_18963672_20230320_065207_EER_patch_aligned [png] [pdf]

Power Spectrum for 007658629387853597550_FoilHole_20039935_Data_18963670_18963672_20230320_065207_EER_patch_aligned [png] [pdf]

[CPU: 585.3 MB] – 0.0: processing 9 of 14646: J20/motioncorrected/003392412203048862050_FoilHole_20039935_Data_18963676_18963678_20230320_065215_EER_patch_aligned.mrc loading /mnt/nfs/user_data/Sa_Chen/cryosparc_projects/SC25_1/P27/J20/motioncorrected/003392412203048862050_FoilHole_20039935_Data_18963676_18963678_20230320_065215_EER_patch_aligned.mrc Loading raw mic data from J20/motioncorrected/003392412203048862050_FoilHole_20039935_Data_18963676_18963678_20230320_065215_EER_patch_aligned.mrc … Done in 0.03s Processing … Done in 2.45s Completed patch CTF estimation with (Y:2,X:2) knots
1D search for 000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned [png] [pdf]

1D search result for 000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned [png] [pdf]

2D patch result for 000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned [png] [pdf]

Diagnostic plot for 000408107024480566766_FoilHole_20039935_Data_18963673_18963675_20230320_065211_EER_patch_aligned [png] [pdf]

[CPU: 456.4 MB] Traceback (most recent call last): File “cryosparc_worker/cryosparc_compute/run.py”, line 85, in cryosparc_compute.run.main File “cryosparc_worker/cryosparc_compute/jobs/ctf_estimation/run.py”, line 353, in cryosparc_compute.jobs.ctf_estimation.run.run File “/home/ldbuser/software/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.7/site-packages/matplotlib/pyplot.py”, line 1586, in ylim ret = ax.set_ylim(*args, **kwargs) File “/home/ldbuser/software/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.7/site-packages/matplotlib/axes/_base.py”, line 3568, in set_ylim bottom = self._validate_converted_limits(bottom, self.convert_yunits) File “/home/ldbuser/software/cryosparc/cryosparc_worker/deps/anaconda/envs/cryosparc_worker_env/lib/python3.7/site-packages/matplotlib/axes/_base.py”, line 3214, in _validate_converted_limits raise ValueError(“Axis limits cannot be NaN or Inf”) ValueError: Axis limits cannot be NaN or Inf
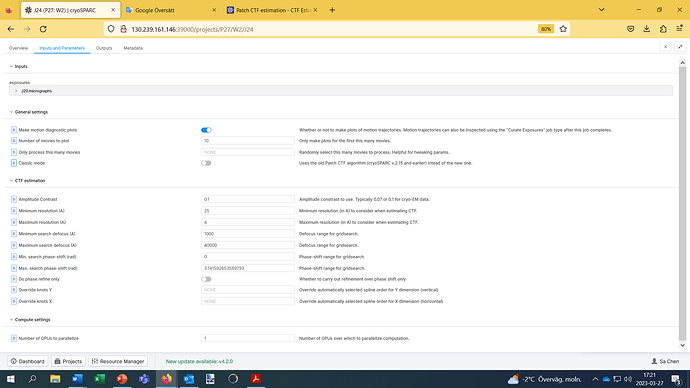
I am not sure if you may see the image (screenshoot). In case it did not work, the parameters are:
make motion diagostic plots On;
Number of moniewto plot 10
Classic mode off
CTF ESTIMATON
Amplitude contrast: 0.1
Minimum resolution (A): 25
Maximum resoluton (A): 4
Minimum search defocus (A): 1000
Maximum searcg deficus (A): 40000
Min.search phase-shift (rad): 0
Max. search phase-shift (rad): 3.1415926…
Computer settings
Number of GPUs to paralielize: 1
Many thanks!
Sa