Hi,
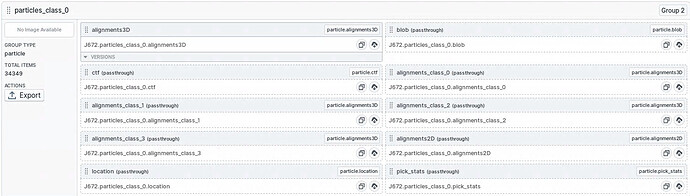
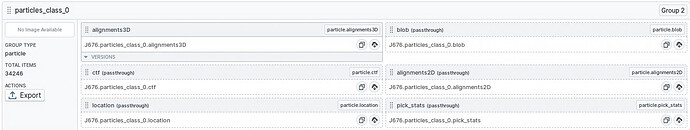
I am trying to figure out some issues with an outside scripting package. When looking at the passthrough outputs for a heterogeneous refinement job, I see that (in this particular case) each class contains the alignment information for all classes. These “extra” alignments are propagating through the passthrough data for downstream NU-refinement jobs. Is this the intended behavior? Other hetero refinement jobs in this workspace do not have the alignments for all classes passed through for each individual class. Its not clear what I have done differently this time.
CryoSPARC v3.0.0 (sorry if this was already patched)
Thanks!