Hi @opornillos and @olibclarke! This is an interesting problem!
Just to make sure I’m following, you have set A and set B, picked from the same micrographs. Your desired result is the set of particles which are in (or, are close to each other in) both A and B, and no other particles.
The problem you note with Oli’s method (remove entirely and use the removed particles) is that the radius you need to use will also create some A/A pairs and B/B pairs, which is undesirable.
If I have all that right, I would definitely reach for CryoSPARC Tools! Here’s an outline of how I might proceed:
First, I’d start with Oli’s method to select particles which are certainly near other particles: that is, all A/A, A/B, and B/B pairs. I would start this way because comparing positions is relatively slow, and this will throw out particles that are definitely not in your final desired set.
Then, working micrograph by micrograph (which you could do using dataset.query() in a for loop over the micrograph UIDs), I would try the following process:
- Get the X,Y coordinates for each particle in A. These are found in the fields
location/center_{x,y}_frac.
- Convert those distances to pixels by multiplying by the relevant
location/micrograph_shape values
- Do the same for particles in B.
- Use
scipy.spatial.distance.cdist() to find the pairwise distance between all points in A and all points in B.
Now, you have an MxN matrix, where M and N are the number of particles in A and B. Each entry (i,j) is the distance between the ith particle from A and the jth particle from B. So, you can follow this procedure to keep only the particles that are some minimum distance away from any particle in A:
- Iterate over the particles in B, with some index j.
- Check the minimum distance of the j-th column in your distance matrix. If it is below your threshold, save the relevant B particle’s UID.
Once that’s done, you’ll have a list of UIDs which belong to B particles that are some distance away from an A particle. With that, a simple query() will get you the set you want, and it can be saved using save_external_job().
Here is some example code starting with a fake list of A and B particle coordinates:
import numpy as np
from scipy.spatial import distance
a_particles = np.array([[0,0],
[1,1],
[0,2],
[1,3]])
b_particles = np.array([[2.0,1.0],
[1,0.3],
[1.1,3.1]])
distances = distance.cdist(a_particles, b_particles)
maximum_distance = 0.8
for j in range(len(b_particles)):
nearest_a_distance = np.min(distances[:,j])
if nearest_a_distance <= maximum_distance:
print(f'Keep UID {j}')
# output
Keep UID 1
Keep UID 2
And here’s a plot representing, graphically, what we’re doing here. The A particles are black points. The B particles are numbered by their “UID”, and have a green circle with radius maximum_distance. If this radius overlaps a black point, we keep the particle. Otherwise, we discard it.
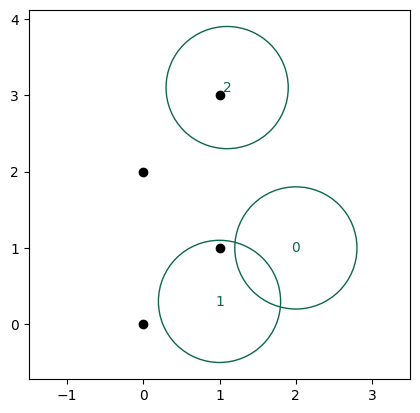
You can see that even though B particles 0 and 1 would be counted as duplicates (their circles overlap), we correctly rejected B particle 0 because it is not close enough to any A particle.
I hope that helps!
