I am utilizing the helical processing tutorial for a nanofiber reconstruction but encountered an error message upon queuing the filament tracing job
RuntimeError: cuda failure (driver API): cuMemAlloc(&plan_cache.plans[idx].workspace, plan_cache.plans[idx].worksz) → CUDA_ERROR_OUT_OF_MEMORY out of memory
I am unsure how to resolve this issue and move forward with the reconstruction
You will need to provide more info - what system are you running this on? which GPU cards with how much VRAM? etc
2 Likes
You can try a stronger lowpass filter, but that isn’t always enough. That CUDA error means you need a GPU with more VRAM, though. But as Oli says, more information (about both system and dataset) is really needed.
1 Like
In relation to the system we have the Dell Inc. Precision 3660 hardware model – the memory is 128.0 GiB – the processor is the 13th Gen Intel Core i7-13700 x 24 – I believe the GPU is NVIDIA RTX A4000 – the disk capacity is 1.0 TB and the operating system we have is Ubuntu 22.04.5 LTS – the operating system type is 64-bit – the GNOME version is 42.9 – the windowing system is X11
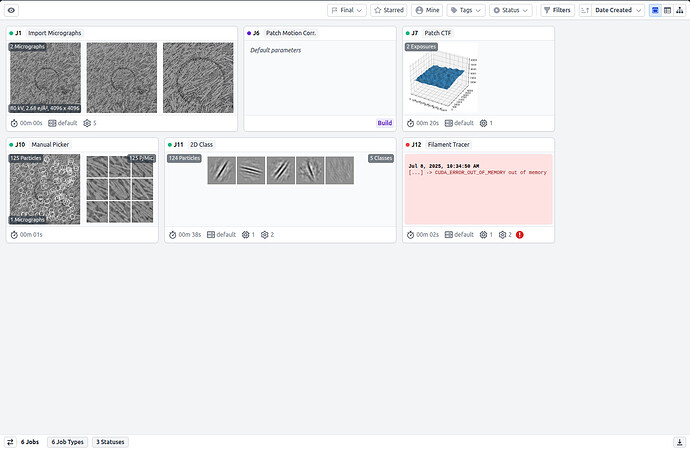
In relation to the dataset it consists of two mrc files I was advised to reconstruct which I imported as micrographs – carried out the Patch CTF – moved forward with the manual picker in which I picked 125 particles – followed by 2D Class to create templates which resulted in 5 templates – dragged and dropped the micrographs picked from the outputs of the manual picker and the class averages from the 2D class into the filament tracer job and input the diameter in A and the separation distance the remaining values were default
I am hopeful the excess information is useful in determining the issue 
@acabrer2 what box sizes are you working with ? over 800pix can push the limits of the A5000s, so not sure about the A4000 or if trying low memory mode will help.
2 Likes
I believe the box size is 4096 x 4096 and the pixel size is 455.3 pm could this push the limits?
That should be the micrograph dimensions and the pixel spacing/sampling frequency/Angstrom per pixel (different words, effectively mean the same thing), respectively, i.e.: 4K micrographs (therefore from Falcon camera, or possibly an Apollo) and 0.455 Angpix.
How large are the boxes you are extracting your 125 picked particles from?
If possible, post a screenshot of your workflow. 
2 Likes
This was probably a Falcon detector, Gatans are usually not square. Either way, as @rbs_sci points out, each micrograph has 4k pixels in one dimension and the extraction box size should be smaller e.g. 324pix (we just assume it is a square so each extracted particle is a 324x324pix .mrc/.mrcs that is then processed in Fourier space. Some virus people that want high resolution will collect at ~0.5A/pix, then only have a few particles per micrograph - so their box sizes can be nearly as big as the micrograph ~1k pix. In general with cryosprc, we have had issues working with particle stacks extracted over 1024pix.
@acabrer2 In your case I would down sample these by “binning” the same idea as “Fourier crop” in the extract job. So in the extract from micrographs job, if your extraction box is 512, keep that but under Fourier Crop ender something like 128 or even 64. When this particle stack is reconstructed or refined you will see the FSC hitting Nyquist frequency (the right side of the plot) because your actual box size will be 4-8x larger than the actual recorded data. So then you can decrease binning or increase Fourier crop (64, 128, => 256) until your system cannot handle it. In many cases with data sets collected at ~0.5A/pix you never really get close to Nyquist (2x pixel size = 1A) so it is faster & less disk space to work with such particle stacks.
*also for the extract job, ensure float 16 is enabled. There is really no reason to work with 32 floating points.
2 Likes