Dear All,
I am quite now in cryoEM and I am trying to interpret images of an enzyme, multidomain, flexible, monomeric, 150Kda, … cannot do worse than that !
I have a question on the ideal number of classes to start an ab-initio reconstruction.
It is better to start with just few classes (5-10) or with several classes (10-20), and subsequently re-classify extensively using 3DClass ?
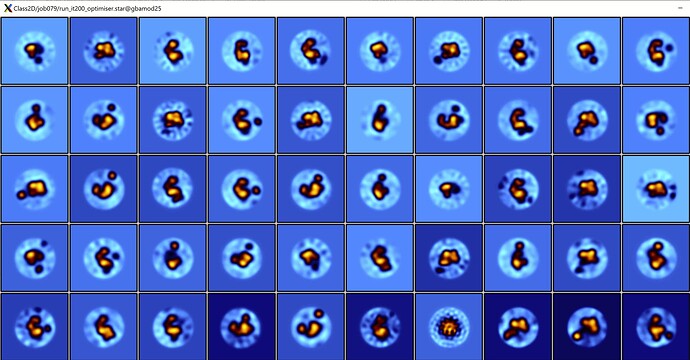
The problem is that my protein does not have a regular shape that I can easily recognize in Class2D, so I actually don’t know which are true “good” and “bad” classes… I might be different orientation, different shape… or both.
I post here some 2Dclasses that I get after a few cycles of “junk cleaning” (I am very sorry the image it’s from Relion but in Cryosparc I obtain identical, if not better, results). I am working now with a set of ~100000 particles.
Thanks a lot for your advice !
Gia