Hi,
I have a dataset from a supposed dimer of 100 kDa. I expect the length of the protein to be about 120 A on the longest side. I’ve been trying to get 2D classes but it didn’t work well. First, I was a bit strict on the blob picker and tried to narrow down the picking to reasonable particles and the 2D classes seemed to be overfitted noise:
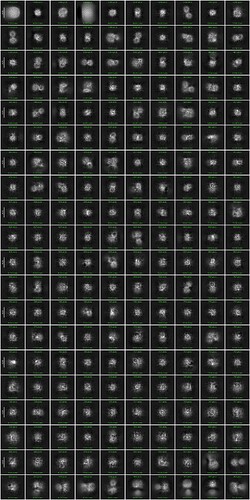
Then I tried 2D classification using much more particles and I did the elliptical blob picking. It didn’t help much.
The 2D classification jobs parameters are default except:
Force max: off
Online EM-iterations: 40
Batchsize: 500
I’d appreciate if anyone can give me some ideas how to approach this. Thanks!