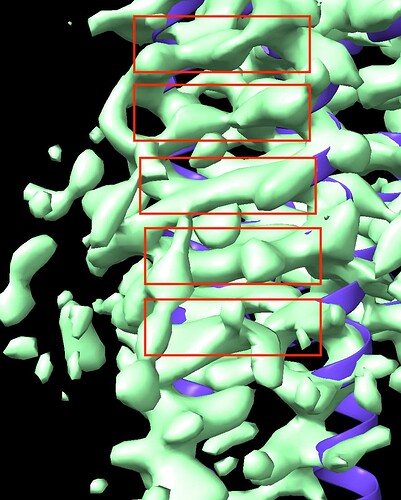
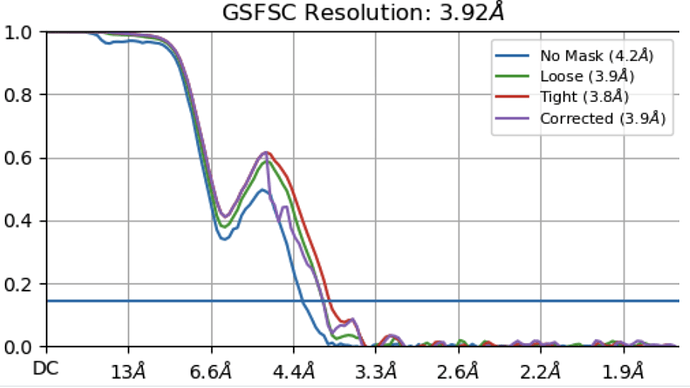
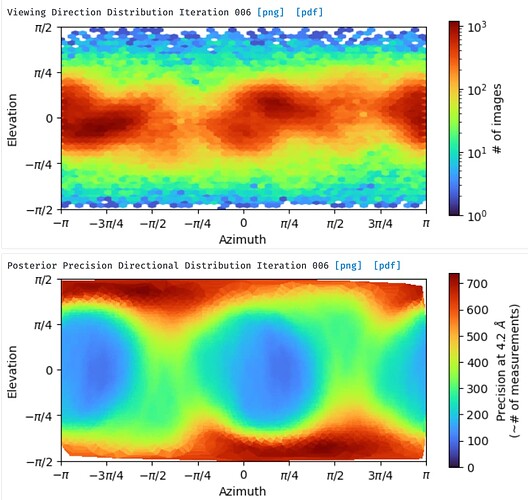
I am characterizing unknown gpcr structure. After local refinement, I got an electron density map which has not bad resolution about late 3. But my map has a point in dispute. Some helix side chains of TM5 and TM6 in my map are so stretched like a spider web or a mozzarella cheese. On the basis of viewing direction distribution map, my map didn’t seem to be preferred orientations. Why so stretched in TM5 and TM6 ? Please reply to me… Thank you and Good Luck for your structure.
Hey @shinjw1887!
Looks like that ‘stretching’ might be the result of a flexing motion. Have you tried running 3D Variability analysis to see if you can associate that flexibility with one or more variability components? If that works, you might be able to cluster your particles to try and remove (some) of that stretching.
Valentin
Good Suggestion. Thank you. I will do what you said.
Agree with @vperetroukhin - we have seen similar anisotropy with interdomain flexibility. The other possibility I would consider, looking at your orientation distribution, is pseudosymmetry - what symmetry are you imposing, and what is the approximate symmetry of your molecule?
I’m imposing C1 symmetry.
My molecule looks like this.
Right - looking at this I would expect that interdomain flexibility is the culprit. I would try performing a local refinement with a mask just around the membrane embedded portion, if you haven’t already done so - this has helped quite a lot in similar cases. 3D-VA after local refinement may also be worth a go, to isolate a subset of particles with better TM density.
Chers
Oli
Thank you for your suggestion…!