Hi@HaoweiZhang,
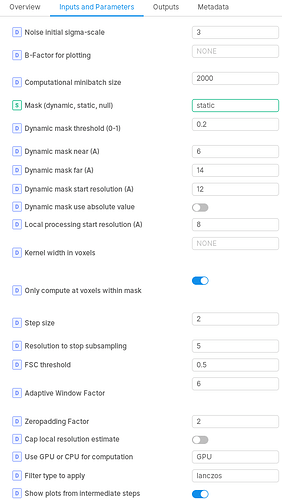
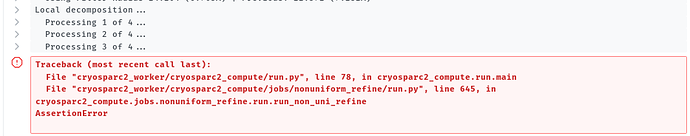
I got a running error below when the NU-refinement tool was used. I have used the default parameters except for change the “Type of masking to use” from dynamic to static since a generated static mask was applied. It stopped after 4 iterations when start local processing, the error message and parameters were listed below.
Thank you.
Best wishes.
when change the “Type of masking to use” option from static to dynamic, there is also a running error appears