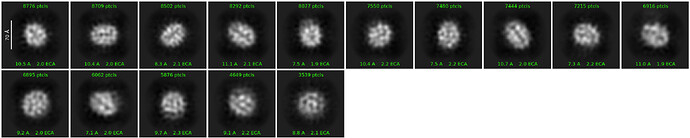
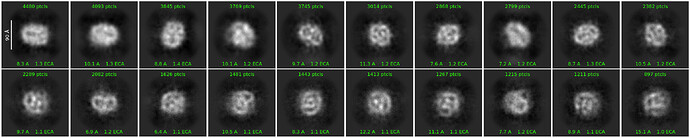
Hello everyone,
I have a nanodisc dataset that is giving me noisy and unrepresentative 2D class averages. I am looking for help to improve the 2D classes. Protein is ~150kDa in an MSP1D1 ND.
I have tried increasing max resolution, increasing minimum separation distance, turning off max poses, increasing batch size, increasing iterations, increasing/decreasing box size. Further suggestions would be very welcome!
Here are some examples of my attempts:
My best run so far has come after curating 18k particles, this has been difficult to reproduce with other sets of particles.
Parameters for best run
5 classes
Max res: 3A
Max alignment res: 3A
Number of iterations: 40
Batchsize: 400
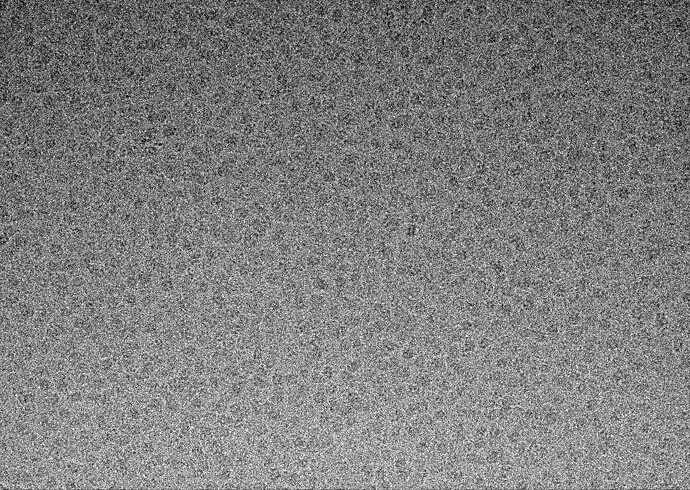
Example Micrograph
Perhaps my problem is upstream of 2D classification?