Hi there,
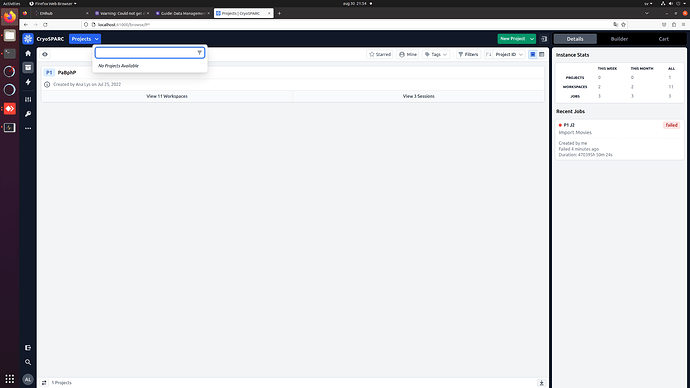
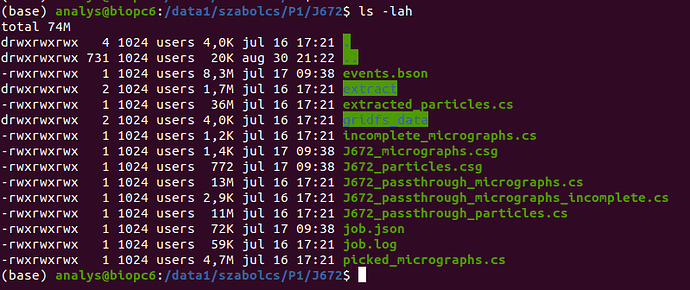
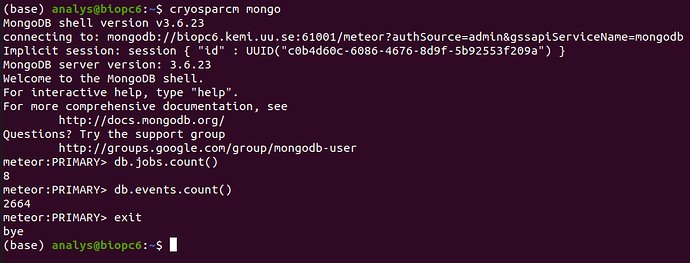
I have recently had a hard disk failure with the disk containing the home directory and the cryosparc database. I have recovered the disk following this guide (recovering the superblock from a backup on the disk), but the cryosparc_database got corrupted. There was no suitable backup of the database (huge mistake, I know), and I was unable to restore it with mongod. I tried to delete the database and creating an empty one following this solution, which seems to have worked. Trying to attach P1 failed due to a live session which was running at the time of the disk failure, so I have deleted the session folder from the project directory. After this, P1 was successfully (?) attached and the workspaces and live sessions appear:
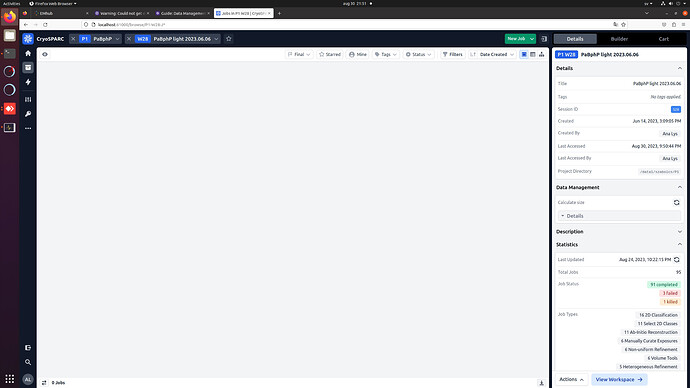
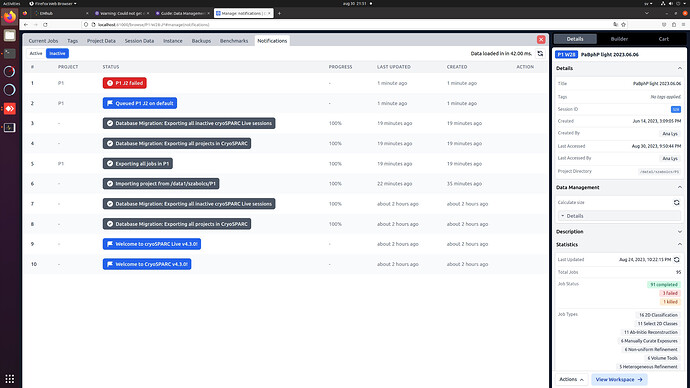
The jobs, however, do not show up within the workspaces, even though the statistics (number of completed/failed/killed jobs) are correctly showing in the workspace details:
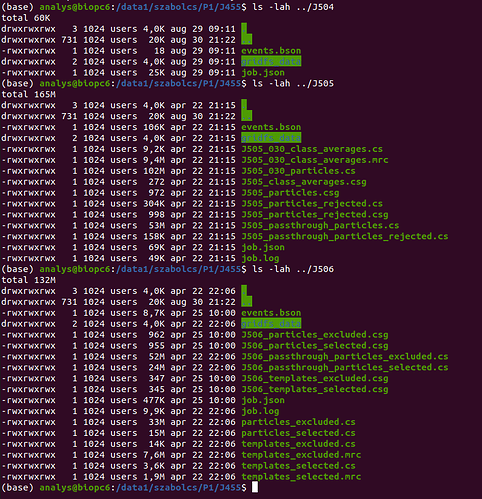
Trying to run any new jobs fails as an existing job ID (starting at J1) is assigned to them. Running cryosparcm test workers also fails for the same reason.
Is there a way to salvage my earlier jobs? I think starting a new project should work for any later data processing, but if possible, it would be nice to get my earlier results back.
Job log from the failed worker test:
================= CRYOSPARCW ======= 2022-07-25 11:01:55.677029 =========
Project P1 Job J1
Master biopc6.bcbp.gu.se Port 61002
===========================================================================
========= monitor process now starting main process
MAINPROCESS PID 14836
MAIN PID 14836
imports.run cryosparc_compute.jobs.jobregister
========= monitor process now waiting for main process
========= sending heartbeat
========= sending heartbeat
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
TIFFReadDirectory: Warning, Bogus "StripByteCounts" field, ignoring and calculating from imagelength.
========= sending heartbeat
TIFFFetchDirectory: Can not read TIFF directory count.
TIFFReadDirectory: Failed to read directory at offset 1572864.
/data1/szabolcs/2022-07-04_Westenhoff_PaPhy-FL-initial_EPU/Images-Disc1/GridSquare_3686077/Data/FoilHole_4773925_Data_3688194_3688196_20220705_190027_Fractions.tif: Not a TIFF or MDI file, bad magic number 0 (0x0).
/data1/szabolcs/2022-07-04_Westenhoff_PaPhy-FL-initial_EPU/Images-Disc1/GridSquare_3686077/Data/FoilHole_4773937_Data_3688194_3688196_20220705_190546_Fractions.tif: Not a TIFF or MDI file, bad magic number 0 (0x0).
***************************************************************
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
========= sending heartbeat
min: 60566.393555 max: 73381.872070
min: 1030643.386719 max: 1112529.113281
min: 1030643.386719 max: 1112529.113281
min: 252635.568848 max: 283566.743652
min: 60659.390869 max: 73373.327881
min: 60553.894531 max: 73431.792969
***************************************************************
Traceback (most recent call last):
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 870, in run_import_movies_or_micrographs
result.get()
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 657, in get
raise self._value
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 121, in worker
result = (True, func(*args, **kwds))
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 788, in header_check_worker
eer_upsamp_factor=eer_upsamp_factor)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 677, in read_movie_header
shape = tiff.read_tiff_shape(abs_path)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/blobio/tiff.py", line 69, in read_tiff_shape
tif = libtiff.TIFF.open(path)
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/site-packages/libtiff/libtiff_ctypes.py", line 501, in open
raise TypeError('Failed to open file ' + repr(filename))
TypeError: Failed to open file b'/data1/szabolcs/2022-07-04_Westenhoff_PaPhy-FL-initial_EPU/Images-Disc1/GridSquare_3686077/Data/FoilHole_4773923_Data_3688194_3688196_20220705_185941_Fractions.tif'
Traceback (most recent call last):
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 870, in run_import_movies_or_micrographs
result.get()
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 657, in get
raise self._value
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 121, in worker
result = (True, func(*args, **kwds))
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 788, in header_check_worker
eer_upsamp_factor=eer_upsamp_factor)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 677, in read_movie_header
shape = tiff.read_tiff_shape(abs_path)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/blobio/tiff.py", line 69, in read_tiff_shape
tif = libtiff.TIFF.open(path)
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/site-packages/libtiff/libtiff_ctypes.py", line 501, in open
raise TypeError('Failed to open file ' + repr(filename))
TypeError: Failed to open file b'/data1/szabolcs/2022-07-04_Westenhoff_PaPhy-FL-initial_EPU/Images-Disc1/GridSquare_3686077/Data/FoilHole_4773925_Data_3688194_3688196_20220705_190027_Fractions.tif'
Traceback (most recent call last):
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 870, in run_import_movies_or_micrographs
result.get()
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 657, in get
raise self._value
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/multiprocessing/pool.py", line 121, in worker
result = (True, func(*args, **kwds))
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 788, in header_check_worker
eer_upsamp_factor=eer_upsamp_factor)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/jobs/imports/run.py", line 677, in read_movie_header
shape = tiff.read_tiff_shape(abs_path)
File "/home/analys/cryosparc/cryosparc_master/cryosparc_compute/blobio/tiff.py", line 69, in read_tiff_shape
tif = libtiff.TIFF.open(path)
File "/home/analys/cryosparc/cryosparc_master/deps/anaconda/envs/cryosparc_master_env/lib/python3.7/site-packages/libtiff/libtiff_ctypes.py", line 501, in open
raise TypeError('Failed to open file ' + repr(filename))
TypeError: Failed to open file b'/data1/szabolcs/2022-07-04_Westenhoff_PaPhy-FL-initial_EPU/Images-Disc1/GridSquare_3686077/Data/FoilHole_4773937_Data_3688194_3688196_20220705_190546_Fractions.tif'
========= main process now complete.
========= monitor process now complete.