Hi everyone!
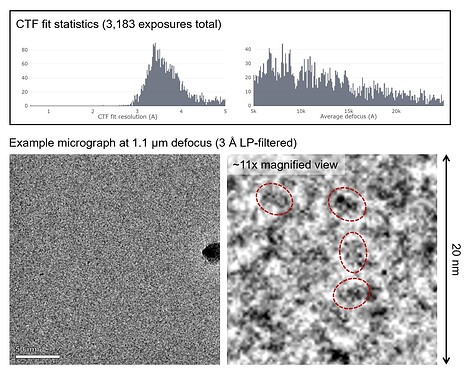
I’m working on the structure of a small asymmetric complex (~60 kDa). The particles are visible with reasonable contrast but are quite crowded. The high particle density was a deliberate choice as it helped with accurate CTF estimation (by providing more signal) as well as attaining thin ice <20 nm.
Fig. 1. CTF fit statistics and a representative micrograph with a few example particles circled
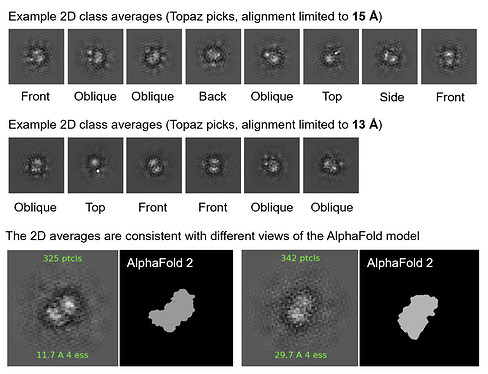
I extracted ~1.6 M Topaz-picked particles and was able to get good-looking 2D classes that were consistent with the expected structure of the complex, though the resolution was not good enough to resolve secondary structure elements. I was quite satisfied with the 2D results given the small size of the target complex in question.
Fig. 2. Representative 2D class averages from the Topaz-picked particles (~1.6 M)
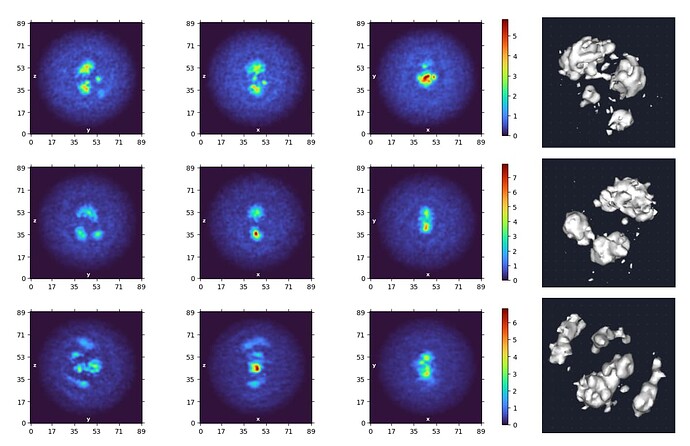
However, when I moved onto ab initio, I found that regardless of the parameters used (real-space windowing, # of classes, etc.) the algorithm would consistently generate volumes with more than one particle in view. The off-center particles throw off the alignment and lead to blurring/distortion of the target particle in the center. This is not too surprising as hints of nearby particles are visible even in the 2D averages (presumably less affected than ab initio due to fewer angular parameters that need to be aligned).
Fig. 3. Results from a 3-class ab initio run using a 2D-curated particle stack
Does anyone have any suggestions as to how this issue may be addressed? Any comments or suggestions are greatly appreciated!
Thanks!
Sincerely,
Joonyoung
trim your box size for extraction, around 120% size of your longest version
1 Like
Dear @ccgauvin94 and @V3eneno,
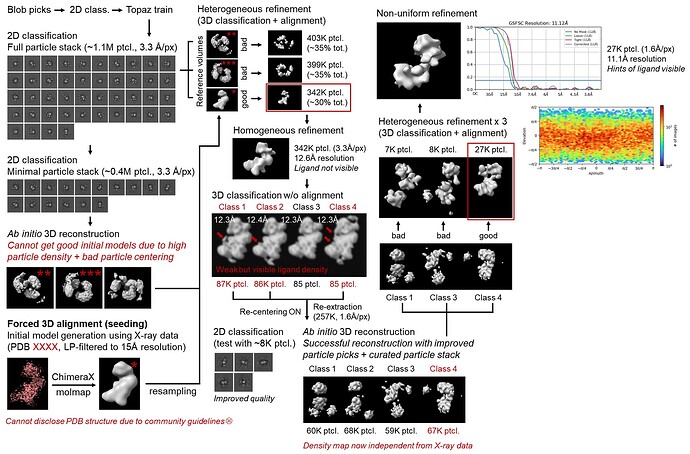
Thanks a lot for your suggestions! I have tried all of them out - (1) reducing box size to ~120% of the expected particle size, (2) masking out + re-centering all but one of the particles from the dirty ab initio volumes before 3D refinement, (3) increasing the dynamic masking threshold during 3D refinement, (4) and using an external reference volume for initial 3D refinements.
What ended up working was option #4, where I used a 15 Å LP-filtered map generated from a known X-ray structure of the apo form of my protein (my sample is the same protein bound to an oligonucleotide ligand) for initial refinements. Specifically, I used hetero refine to first sort out particles that align well to the reference map and used homo refine + 3D classification to get rid of overfitting as much as possible.
When I re-extracted the particles with recenter using aligned shifts ON, the particle picks were much better centered than the initial Topaz picks and allowed me to obtain a good ab initio volume (without the multi-particle issue) straight from my Krios data alone. It seems that the particle picks were the main cause of the issue after all. This is certainly progress, and I’m grateful to both of you for your kind suggestions 
(Sorry I had to blur some of the images due to community guidelines)
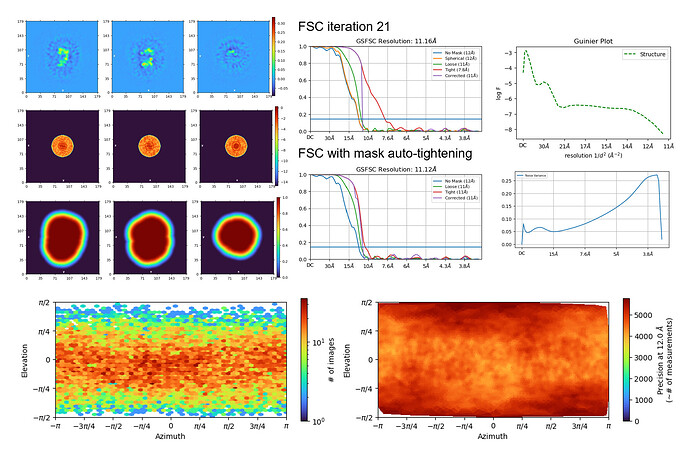
However, I’m still not able to achieve <10Å resolution even after rounds of extensive particle curation. I guess I could re-train Topaz using the new centered particle picks and extract again to make my particle stack bigger (since I only have ~27K particles left), but I don’t think that alone would solve the 10Å barrier. Below are some of the diagnostic plots from my final NU-refine run that hit 11.1 Å resolution.
The algorithm doesn’t overfit too much (i.e. generate spikes all over the place) regardless of alignment resolution or # of iterations, but the resolution doesn’t improve too much either. I understand that this is a very challenging particle to align to high resolution, but I believe it should be possible based on the quality of the micrographs. The fact that the algorithm doesn’t overfit also tells me that the particle stack is quite clean.
Does anyone have any suggestions as to what I could try to improve resolution in a case like this?
Thanks as always!
Sincerely,
Joonyoung
1 Like