Hi
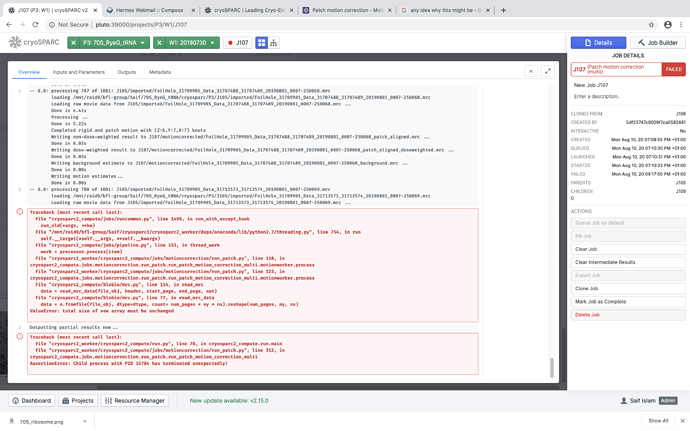
I am facing an issue with Motion Correction. It gets stopped exactly after 787th movie with the follow error. Any help regarding how to resolve the issue will be greatly appreciated.
Traceback (most recent call last):
File “cryosparc2_compute/jobs/runcommon.py”, line 1490, in run_with_except_hook
run_old(*args, **kw)
File “/mnt/raid0/bfl-group/Saif/cryosparc3/cryosparc2_worker/deps/anaconda/lib/python2.7/threading.py”, line 754, in run
self.__target(*self.__args, **self.__kwargs)
File “cryosparc2_compute/jobs/pipeline.py”, line 153, in thread_work
work = processor.process(item)
File “cryosparc2_worker/cryosparc2_compute/jobs/motioncorrection/run_patch.py”, line 118, in cryosparc2_compute.jobs.motioncorrection.run_patch.run_patch_motion_correction_multi.motionworker.process
File “cryosparc2_worker/cryosparc2_compute/jobs/motioncorrection/run_patch.py”, line 123, in cryosparc2_compute.jobs.motioncorrection.run_patch.run_patch_motion_correction_multi.motionworker.process
File “cryosparc2_compute/blobio/mrc.py”, line 114, in read_mrc
data = read_mrc_data(file_obj, header, start_page, end_page, out)
File “cryosparc2_compute/blobio/mrc.py”, line 77, in read_mrc_data
data = n.fromfile(file_obj, dtype=dtype, count= num_pages * ny * nx).reshape(num_pages, ny, nx)
ValueError: total size of new array must be unchanged
Outputting partial results now…
Traceback (most recent call last):
File “cryosparc2_worker/cryosparc2_compute/run.py”, line 78, in cryosparc2_compute.run.main
File “cryosparc2_worker/cryosparc2_compute/jobs/motioncorrection/run_patch.py”, line 312, in cryosparc2_compute.jobs.motioncorrection.run_patch.run_patch_motion_correction_multi
AssertionError: Child process with PID 15784 has terminated unexpectedly!
Many thanks in advance
Saif