Hi,
Would it be possible after signal subtraction to keep linked copies of the original particles as a separate blob?
This would be useful in the case that one selects a subset of particles after signal subtraction, and then wants to go back and perform a refinement or classification on the original particles, but using just the subset selected after signal subtraction.
Cheers
Oli
1 Like
Can you already do this by expanding the input particles and replacing their blob with the blob from the output tab of the other particles?
Not as far as I can tell but entirely possible I’m missing something
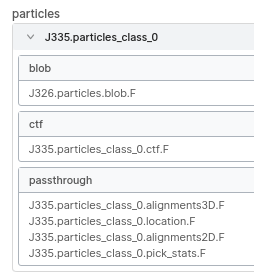
You can click the arrow next to the particles after adding them as input, and it will expand to show you the data associated with those particles. If you then also open a different job and go to the outputs tab, you can drag the particles.blob from the output to the one in the expanded input particles, and it will be replaced. It will get highlighted in green to indicate it’s different. Here’s a screenshot of the input parameters from a finished job where I did that (to use a classification selection from binned particles with the unbinned extraction I had already done). Note the blob is from a different job.
Apologies if you already know this, I haven’t tried for the subtracted->original case.
1 Like
Yes - I know how to do that - but I don’t think that helps in this case? Because when the subtracted particles are created, there is no link to identify how they correspond to the original particles, as far as I can tell?
Here’s a script I call npinfo.py, I use it to dump info for .cs files mostly. I don’t have a job ready to check, but maybe there’s actually something in there already.
#!/usr/bin/env python
import numpy as np
import sys
a = np.load(sys.argv[1])
print(a.shape)
print(a.dtype.names)
If not, then IMO it would be best to enable the blob switch for the subtracted particles, at most it’s one additional piece of information to store (original particle UID) per subtracted particle. On a case by case basis you could do it with csparc2star.py and the merging functionality in star.py, but it is pretty annoying.
1 Like
Hi @olibclarke
@DanielAsarnow is correct - the linkage between particles before/after signal subtraction is maintained in the .cs files, in the form of the uid column. The uid of a given particle is fixed at extraction time, and that serves as a unique stamp for all related versions of that particle (eg. after downsampling, subtraction, etc). So if you connect the subset you have after subtraction to the next job in your workflow, but over-ride the blob using low level results as @DanielAsarnow said, that next job will use the subset you want but load the original un-subtracted blobs.
1 Like
OK I will give it a go! I still think it would be better to have a more accessible way of accessing the matching original particles - if this works then great, but why not just have the unsubtracted particles present as a separate blob in the higher level results section?
OK so I just tried this and it does work - I am a little confused as to how though.
I start a new refinement job from the subtracted particles.
In low level results, it has a particles_selected blob, a particles_selected.ctf blob, and alignments. I replace the particles_selected blob with one from an earlier refinement of unsubtracted particles, and that seems to work - it selects the right number of particles - but where is it getting the particle selection from now that I have replaced the particles blob? From particles_selected.CTF?
Apologies for being dim
Cheers
Oli
I believe the particles.blob is just a mongodb document store for the corresponding particle .cs file. The keys are the particle UIDs, which can be used for a database merge between any table of particles within the same project (or workspace?). If you override part of an input and the UIDs agree, then it does the join and pulls in the relevant fields (the ones within the “blob” you replaced). Offsets are stored in Angstroms, and the dimension and pixel size are also stored in each .cs file, so the alignments & CTFs can easily be set correctly. (Just an educated guess).
Edit
If that’s unclear - when you input some particles, it starts from that table of particles. Whatever pieces of the input you replace individually are merged via the UID key and the corresponding information (particle stack path, index, pixel size) is used to build the particles table in the new job. FYI, pyem uses a field called ucsfParticleUid internally to simplify conversion and matching of particles, I just couldn’t save it in the final star files because versions of Relion before 3.1 are hard-coded to error out if they see a .star field that isn’t from the pre-approved list.
1 Like
so if there are effectively two overlapping sets of particles, it’ll take the intersection - makes sense
