@wtempel My apologies for the delay in my reply, please find below the output of the 4 commands:
cryosparcm cli "get_job('$cspid', '$csjid', 'job_type', 'version', 'params_spec', 'instance_information', 'input_slot_groups', 'status')"
{'_id': '66a08adab4fd87e3b66cc706', 'input_slot_groups': [{'connections': [{'group_name': 'flex_model', 'job_uid': 'J986', 'slots': [{'group_name': 'flex_model', 'job_uid': 'J986', 'result_name': 'checkpoint', 'result_type': 'flex_model.checkpoint', 'slot_name': 'checkpoint', 'version': 'F'}]}], 'count_max': 1, 'count_min': 1, 'description': '', 'name': 'flex_model', 'repeat_allowed': False, 'slots': [{'description': '', 'name': 'checkpoint', 'optional': False, 'title': 'Checkpoint', 'type': 'flex_model.checkpoint'}], 'title': '3DFlex model', 'type': 'flex_model'}, {'connections': [{'group_name': 'particles', 'job_uid': 'J983', 'slots': [{'group_name': 'particles', 'job_uid': 'J1004', 'result_name': 'blob', 'result_type': 'particle.blob', 'slot_name': 'blob_fullres', 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'ctf', 'result_type': 'particle.ctf', 'slot_name': 'ctf', 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'alignments3D', 'result_type': 'particle.alignments3D', 'slot_name': 'alignments3D', 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'blob_train', 'result_type': 'particle.blob', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'blob_train_ctf', 'result_type': 'particle.blob', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'blob', 'result_type': 'particle.blob', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'alignments2D', 'result_type': 'particle.alignments2D', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'filament', 'result_type': 'particle.filament', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'pick_stats', 'result_type': 'particle.pick_stats', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'location', 'result_type': 'particle.location', 'slot_name': None, 'version': 'F'}, {'group_name': 'particles', 'job_uid': 'J983', 'result_name': 'sym_expand', 'result_type': 'particle.sym_expand', 'slot_name': None, 'version': 'F'}]}], 'count_max': 1, 'count_min': 1, 'description': 'Particle stacks to use. Multiple stacks will be concatenated.', 'name': 'particles', 'repeat_allowed': False, 'slots': [{'description': '', 'name': 'blob_fullres', 'optional': False, 'title': 'Particle data blobs', 'type': 'particle.blob'}, {'description': '', 'name': 'ctf', 'optional': False, 'title': 'Particle ctf parameters', 'type': 'particle.ctf'}, {'description': '', 'name': 'alignments3D', 'optional': False, 'title': 'Particle 3D alignments', 'type': 'particle.alignments3D'}], 'title': 'Prepared particles', 'type': 'particle'}], 'instance_information': {'CUDA_version': '11.8', 'available_memory': '1.90TB', 'cpu_model': 'AMD EPYC 7763 64-Core Processor', 'driver_version': '12.2', 'gpu_info': [{'id': 0, 'mem': 84987740160, 'name': 'NVIDIA A100-SXM4-80GB'}], 'ofd_hard_limit': 131072, 'ofd_soft_limit': 1024, 'physical_cores': 128, 'platform_architecture': 'x86_64', 'platform_node': 'saion-gpu26.oist.jp', 'platform_release': '4.18.0-477.15.1.el8_8.x86_64', 'platform_version': '#1 SMP Wed Jun 28 15:04:18 UTC 2023', 'total_memory': '1.97TB', 'used_memory': '59.24GB'}, 'job_type': 'flex_highres', 'params_spec': {'flex_do_noflex_recon': {'value': False}, 'scheduler_no_check_inputs_ready': {'value': True}}, 'project_uid': 'P3', 'status': 'failed', 'uid': 'J1005', 'version': 'v4.4.1'}
cryosparcm eventlog "$cspid" "$csjid" | head -n 40
License is valid.
Launching job on lane largegpu target largegpu ...
Launching job on cluster largegpu
====================== Cluster submission script: ========================
==========================================================================
#!/usr/bin/env bash
#### cryoSPARC cluster submission script template for SLURM
## Available variables:
## /path/to/cryosparc_worker/bin/cryosparcw run --project P3 --job J1005 --master_hostname hostname --master_command_core_port 39002 > /path/to/project/folder/J1005/job.log 2>&1 - the complete command string to run the job
## 4 - the number of CPUs needed
## 1 - the number of GPUs needed.
## Note: the code will use this many GPUs starting from dev id 0
## the cluster scheduler or this script have the responsibility
## of setting CUDA_VISIBLE_DEVICES so that the job code ends up
## using the correct cluster-allocated GPUs.
## 64.0 - the amount of RAM needed in GB
## /path/to/project/folder/J1005 - absolute path to the job directory
## /path/to/project/folder - absolute path to the project dir
## /path/to/project/folder/J1005/job.log - absolute path to the log file for the job
## /path/to/cryosparc_worker/bin/cryosparcw - absolute path to the cryosparc worker command
## --project P3 --job J1005 --master_hostname saion-login1.oist.jp --master_command_core_port 39002 - arguments to be passed to cryosparcw run
## P3 - uid of the project
## J1005 - uid of the job
## name_of_user - name of the user that created the job (may contain spaces)
## user@email.com - cryosparc username of the user that created the job (usually an email)
##
## What follows is a simple SLURM script:
#SBATCH --job-name cryosparc_P3_J1005
#SBATCH -n 4
#SBATCH --gres=gpu:1
#SBATCH --partition=largegpu
#SBATCH --mem=499G
##SBATCH --mem=300000MB
#SBATCH --output=/path/to/project/folder/J1005/job.log
#SBATCH --error=/path/to/project/folder/J1005/job.log
##available_devs=""
##for devidx in $(seq 0 15);
cryosparcm eventlog "$cspid" "$csjid" | tail -n 40
[CPU RAM used: 443 MB] ***************************************************************
[CPU RAM used: 452 MB] ====== 3D Flex Load Checkpoint =======
[CPU RAM used: 452 MB] Loading checkpoint from J986/J986_train_checkpoint_006600.tar ...
[CPU RAM used: 791 MB] Initializing torch..
[CPU RAM used: 791 MB] Initializing model from checkpoint...
Input tetramesh
[CPU RAM used: 1004 MB] Upscaling deformation model to match input volume size...
Upsampled mask
Upsampled tetramesh
[CPU RAM used: 2877 MB] ====== Load particle data =======
[CPU RAM used: 2878 MB] Reading in all particle data on the fly from files...
[CPU RAM used: 2878 MB] Loading a ParticleStack with 12000 items...
[CPU RAM used: 2878 MB] SSD cache : cache successfully synced in_use
[CPU RAM used: 2878 MB] SSD cache : cache successfully synced, found 32,029.75 MB of files on SSD.
[CPU RAM used: 2878 MB] SSD cache : cache successfully requested to check 12 files.
[CPU RAM used: 2878 MB] Detected file change due to modification time.
[CPU RAM used: 2878 MB] SSD cache : cache requires 8,943 MB more on the SSD for files to be downloaded.
[CPU RAM used: 2878 MB] SSD cache : cache has enough available space.
[CPU RAM used: 2878 MB] Needed | 8,943.07 MB
Available | 10,875,058.64 MB
Disk size | 10,984,866.00 MB
Usable space | 10,974,866.00 MB (reserve 10,000 MB)
[CPU RAM used: 2878 MB] Transferring across 2 threads: batch_000010_downsample.mrc (12/12)
Progress | 8,943 MB (100.00%)
Total | 8,943 MB
Average speed | 6,382.42 MB/s
ETA | 0h 0m 0s
[CPU RAM used: 2879 MB] SSD cache : complete, all requested files are available on SSD.
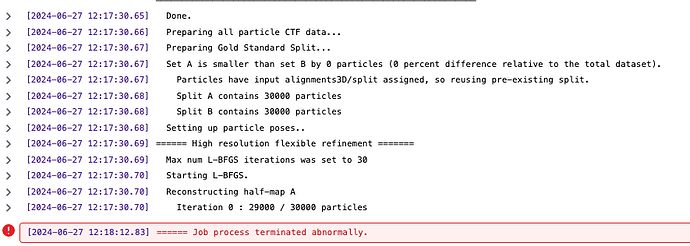
[CPU RAM used: 2884 MB] Done.
[CPU RAM used: 2884 MB] Preparing all particle CTF data...
[CPU RAM used: 2884 MB] Parameter "Force re-do GS split" was off. Using input split..
[CPU RAM used: 2884 MB] Split A contains 6000 particles
[CPU RAM used: 2884 MB] Split B contains 6000 particles
[CPU RAM used: 2884 MB] Setting up particle poses..
[CPU RAM used: 2884 MB] ====== High resolution flexible refinement =======
[CPU RAM used: 2884 MB] Max num L-BFGS iterations was set to 20
[CPU RAM used: 2884 MB] Starting L-BFGS.
[CPU RAM used: 2884 MB] Reconstructing half-map A
[CPU RAM used: 2884 MB] Iteration 0 : 5000 / 6000 particles
[CPU RAM used: 196 MB] ====== Job process terminated abnormally.
cryosparcm joblog "$cspid" "$csjid" | tail -n 40
Project P3 Job J1005
Master master_hostname Port 39002
===========================================================================
========= monitor process now starting main process at 2024-07-24 14:45:33.698634
MAINPROCESS PID 686645
MAIN PID 686645
flex_refine.run_highres cryosparc_compute.jobs.jobregister
========= monitor process now waiting for main process
========= sending heartbeat at 2024-07-24 14:45:48.301647
========= sending heartbeat at 2024-07-24 14:45:58.321484
========= sending heartbeat at 2024-07-24 14:46:08.341454
========= sending heartbeat at 2024-07-24 14:46:18.353252
========= sending heartbeat at 2024-07-24 14:46:28.367796
========= sending heartbeat at 2024-07-24 14:46:38.380905
========= sending heartbeat at 2024-07-24 14:46:48.401071
========= sending heartbeat at 2024-07-24 14:46:58.421064
========= sending heartbeat at 2024-07-24 14:47:08.441066
***************************************************************
Running job J1005 of type flex_highres
Running job on hostname %s largegpu
Allocated Resources : {'fixed': {'SSD': False}, 'hostname': 'largegpu', 'lane': 'largegpu', 'lane_type': 'cluster', 'license': True, 'licenses_acquired': 1, 'slots': {'CPU': [0, 1, 2, 3], 'GPU': [0], 'RAM': [0, 1, 2, 3, 4, 5, 6, 7]}, 'target': {'cache_path': '/scratch', 'cache_quota_mb': None, 'cache_reserve_mb': 10000, 'custom_var_names': ['ram_gb_multiplier'], 'custom_vars': {}, 'desc': None, 'hostname': 'largegpu', 'lane': 'largegpu', 'name': 'largegpu', 'qdel_cmd_tpl': 'scancel {{ cluster_job_id }}', 'qinfo_cmd_tpl': 'sinfo', 'qstat_cmd_tpl': 'squeue -j {{ cluster_job_id }}', 'qstat_code_cmd_tpl': 'squeue -j {{ cluster_job_id }} --format=%T | sed -n 2p', 'qsub_cmd_tpl': 'sbatch {{ script_path_abs }}', 'script_tpl': '#!/usr/bin/env bash\n#### cryoSPARC cluster submission script template for SLURM\n## Available variables:\n## {{ run_cmd }} - the complete command string to run the job\n## {{ num_cpu }} - the number of CPUs needed\n## {{ num_gpu }} - the number of GPUs needed. \n## Note: the code will use this many GPUs starting from dev id 0\n## the cluster scheduler or this script have the responsibility\n## of setting CUDA_VISIBLE_DEVICES so that the job code ends up\n## using the correct cluster-allocated GPUs.\n## {{ ram_gb }} - the amount of RAM needed in GB\n## {{ job_dir_abs }} - absolute path to the job directory\n## {{ project_dir_abs }} - absolute path to the project dir\n## {{ job_log_path_abs }} - absolute path to the log file for the job\n## {{ worker_bin_path }} - absolute path to the cryosparc worker command\n## {{ run_args }} - arguments to be passed to cryosparcw run\n## {{ project_uid }} - uid of the project\n## {{ job_uid }} - uid of the job\n## {{ job_creator }} - name of the user that created the job (may contain spaces)\n## {{ cryosparc_username }} - cryosparc username of the user that created the job (usually an email)\n##\n## What follows is a simple SLURM script:\n\n#SBATCH --job-name cryosparc_{{ project_uid }}_{{ job_uid }}\n#SBATCH -n {{ num_cpu }}\n#SBATCH --gres=gpu:{{ num_gpu }}\n#SBATCH --partition=largegpu\n#SBATCH --mem={{ (ram_gb|float * (ram_gb_multiplier|default(1))|float)|int}}G\n##SBATCH --mem={{ (300*1000)|int }}MB\n#SBATCH --output={{ job_log_path_abs }}\n#SBATCH --error={{ job_log_path_abs }}\n\n\n##available_devs=""\n##for devidx in $(seq 0 15);\n##do\n## if [[ -z $(nvidia-smi -i $devidx --query-compute-apps=pid --format=csv,noheader) ]] ; then\n## if [[ -z "$available_devs" ]] ; then\n## available_devs=$devidx\n## else\n## available_devs=$available_devs,$devidx\n## fi\n## fi\n##done\n##export CUDA_VISIBLE_DEVICES=$available_devs\n\n{{ run_cmd }}\n', 'send_cmd_tpl': '{{ command }}', 'title': 'largegpu', 'tpl_vars': ['num_cpu', 'job_log_path_abs', 'command', 'cluster_job_id', 'ram_gb', 'num_gpu', 'run_cmd', 'run_args', 'project_uid', 'worker_bin_path', 'job_creator', 'project_dir_abs', 'ram_gb_multiplier', 'cryosparc_username', 'job_uid', 'job_dir_abs'], 'type': 'cluster', 'worker_bin_path': '/path/to/cryosparc_saion-login1/cryosparc_worker/bin/cryosparcw'}}
Transferring across 2 threads: (0/12)
Transferring across 2 threads: batch_000001_downsample.mrc (1/12)
Transferring across 2 threads: batch_000000_downsample.mrc (2/12)
Transferring across 2 threads: batch_000003_downsample.mrc (3/12)
Transferring across 2 threads: batch_000002_downsample.mrc (4/12)
Transferring across 2 threads: batch_000004_downsample.mrc (5/12)
Transferring across 2 threads: batch_000005_downsample.mrc (6/12)
Transferring across 2 threads: batch_000006_downsample.mrc (7/12)
Transferring across 2 threads: batch_000007_downsample.mrc (8/12)
Transferring across 2 threads: batch_000008_downsample.mrc (9/12)
Transferring across 2 threads: batch_000009_downsample.mrc (10/12)
Transferring across 2 threads: batch_000011_downsample.mrc (11/12)
Transferring across 2 threads: batch_000010_downsample.mrc (12/12)
========= sending heartbeat at 2024-07-24 14:47:18.455909
========= sending heartbeat at 2024-07-24 14:47:28.468209
========= sending heartbeat at 2024-07-24 14:47:38.488464
========= sending heartbeat at 2024-07-24 14:47:48.507843
========= main process now complete at 2024-07-24 14:47:55.151271.
========= monitor process now complete at 2024-07-24 14:47:55.230972.