Dear colleagues,
We are reconstructing a large oligomeric assembly that forms rings, and encountered trouble with 2D and 3D classification.
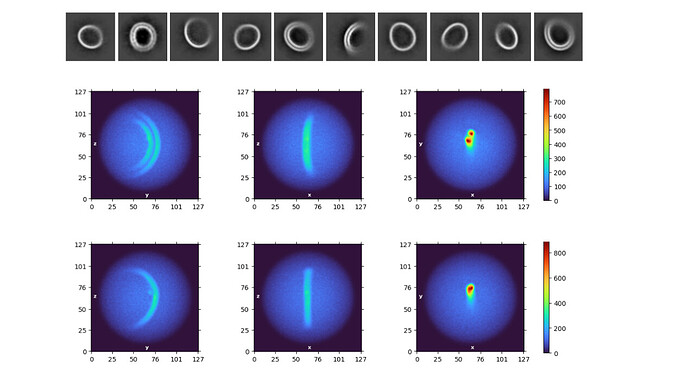
For 2D classification, the rings are often not centered–it seems that for some tilted views, cryoSPARC is centering the apparent center of density, rather than true center of mass, causing the rings to be misaligned (see figure). We get slightly better centering by checking “re-center mask binary”, but this is an imperfect solution.
For ab-initio modelling with C1 symmetry, we get half-rings (see figure)–we speculate that the algorithm cannot distinguish between two sides of the circle when aligning from 2D, but we are uncertain how to proceed. We’ve tried estimating and applying symmetry, but the density becomes very artificial.
Has anyone encountered a similar issue or have tips to overcome these problems with 2D and 3D classification? Thank you!
Environment
cryoSPARC version 3.1
OS Ubuntu