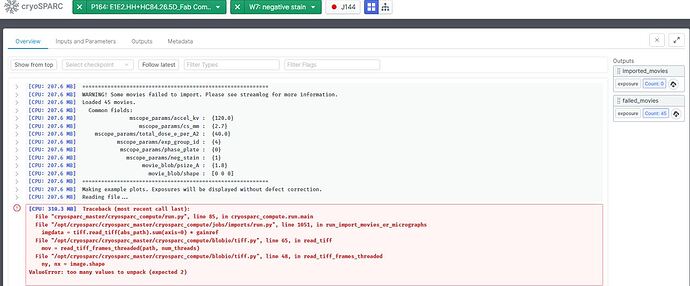
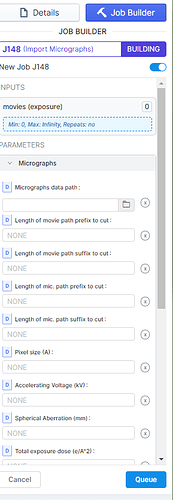
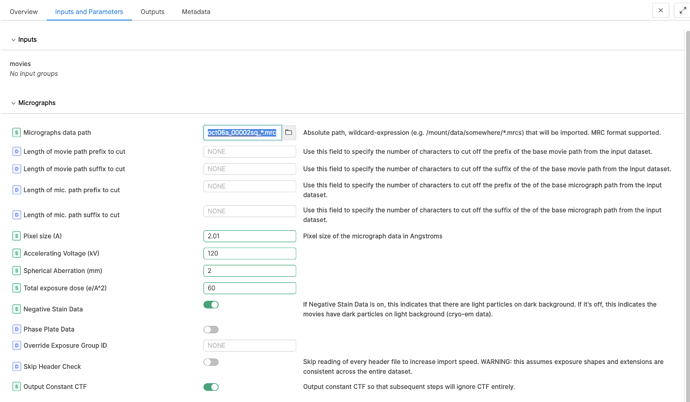
Hello! We recently got some negative staining data from Ceta, not Falcon. Therefore the EER settings don’t apply when I tried to import the data. I tried all different values to get around with this, but the micrographs (tiff files) still failed to import. Here is the error message from the output. Does anyone know how to overcome this? Can I use CryoSPARC for Ceta negative staining data? Is Relion program capable of processing this kind of data (1 set as EMD file, the other set as tiff file)?
Thanks for helping!
Daisy
I think you want import micrographs, not movies, could that be the issue? We have imported NS data from a Ceta before IIRC
Thanks for your response! I wonder how I could import just micrographs in Tiff format. This is the interphase when I try to “import micrographs”, then I have no input in the box of movies. Would you mind showing me the steps how to input just tiff files?
Have you tried selecting the path in the “Micrograph data path” box? Perhaps this doesn’t support TIFF…?
Yes, I did try to put the micrograph path in, but the micrographs have to link to movies (mrc). I don’t have mrc files. What I have are Tiff files and EMD files. I looked at publications, some used Dogpicker or Relion instead of CryoSPARC. I don’t have those programs yet, and just wonder whether CryoSPARC could allow process the data. Thanks for any insight!
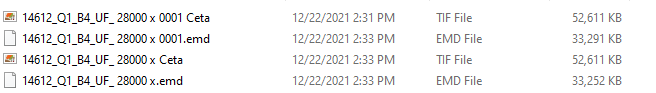
You definitely don’t need to link to movies. We have imported NS data as MRC (see below) - don’t think we have tried importing tiffs. Maybe you can convert tiff to mrc first using IMOD or similar?
(also it is best to first invert the contrast to black on white first, e.g. using relion_image_handler - picking doesn’t work well for NS data directly in cryosparc in my experience)
Okay. I will try IMOD to convert the files.
How to use relion_image_handler? Do I need to download Relion software? Thank you for your advice again!
yes, relion_image_handler is part of relion. If you run it without arguments it will print the various options.
Hope this helps:
To convert multiple tif files to mrc files, all at once:
Install IMOD on your system. Open a bash terminal in the folder in which you have the tif files or cd there. Run the following line in your terminal:
![]()
Here is a bash script to perform tif2mrc on data with a subfolder structure:
#!/bin/bash
# Specify the source directory where your TIFF files are located
src_dir="ADRESS OF THE SOURCE FOLDER"
# Specify the destination directory for MRC files
dest_dir="DESTINATION FOLER"
# Iterate over all .tif files in subdirectories
find "$src_dir" -type f -name "*.tif" | while read -r tif_file; do
# Determine the output directory within the MRC directory
relative_path="${tif_file#$src_dir}"
output_dir="$dest_dir${relative_path%/*}"
mkdir -p "$output_dir"
# Generate the MRC filename by replacing .tif with .mrc
mrc_file="$output_dir/$(basename "${tif_file%.tif}.mrc")"
# Convert the TIFF file to MRC format
tif2mrc -B 0 "$tif_file" "$mrc_file"
done