Hi @lizellelubbe,
Just another thought - I wonder if the flexibility between the two domains is leading to incomplete particle subtraction (because the orientation of each domain with respect to the reference that you are projecting and subtracting varies from image to image).
Perhaps you might try the following:
-
After NU-refine, perform local refinements of each domain (let’s call them domain A and domain B).
-
Perform particle subtraction for domain A, using the particle set from local refinement. Hopefully, subtraction will be more complete, because you are aligning on the region you are going to subtract. However, the domain you now want to refine is now blurred out, with fairly bad starting angles, so let’s fix that:
-
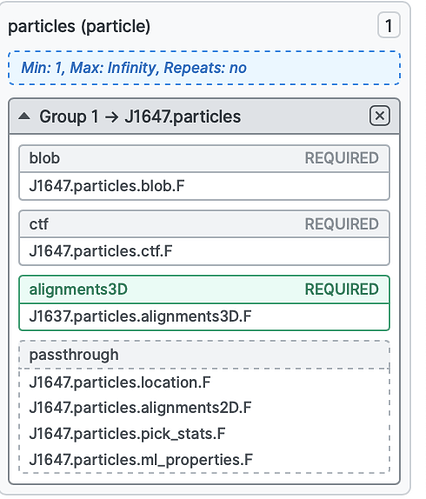
Perform local refinement using the particles from step 2, but the orientations from NU refine, with a mask around the domain you have not subtracted. You can supply the orientations by replacing the low level results group
alignments3Dwith the one from NU-refine, using the low level results interface (see: https://guide.cryosparc.com/processing-data/tutorials-and-case-studies/job-builder-tutorial#fine-tuned-control-over-individual-results). Basically drag thealignments3Dresult group in the output section of the NU-refine job onto the corresponding slot in the job builder for your local refinement job (you will neeed to expand the particles group to see it by clicking the little down arrow):