Hello everyone,
I have a set of simulated particles that I would like to import into CryoSPARC. The particles are in .mrc format, and I learned that I need a .star file to make this possible. Here are the steps I have taken:
- Imported all particles as micrographs.
- Created a custom .star file using cryosparc-tools.
- Used the Import Particles job in CryoSPARC.
I have managed to come a long way, but I am stuck at a point. For some reason, I keep getting this error:
Traceback (most recent call last):
File "cryosparc_master/cryosparc_compute/run.py", line 115, in cryosparc_master.cryosparc_compute.run.main
File "/em_data/localuser/cryoSPARC/cryosparc_worker/cryosparc_compute/jobs/imports/run.py", line 535, in run_import_particles
fig = plotutil.plot_images_simple(dispparts, rows=4, cols=8, radwn=6, figscale=1.5)
File "/em_data/localuser/cryoSPARC/cryosparc_worker/cryosparc_compute/plotutil.py", line 860, in plot_images_simple
rD = img.get_original_real_data()
File "/em_data/localuser/cryoSPARC/cryosparc_worker/cryosparc_compute/particles.py", line 34, in get_original_real_data
data = self.blob.view()
File "/em_data/localuser/cryoSPARC/cryosparc_worker/cryosparc_compute/blobio/mrc.py", line 145, in view
return self.get()
File "/em_data/localuser/cryoSPARC/cryosparc_worker/cryosparc_compute/blobio/mrc.py", line 140, in get
_, data, total_time = prefetch.synchronous_native_read(self.fname, idx_start = self.page, idx_limit = self.page+1)
File "cryosparc_master/cryosparc_compute/blobio/prefetch.py", line 82, in cryosparc_master.cryosparc_compute.blobio.prefetch.synchronous_native_read
OSError:
IO request details:
Error ocurred (Invalid argument) at line 680 in mrc_readmic (1)
The requested frame/particle cannot be accessed. The file may be corrupt, or there may be a mismatch between the file and its associated metadata (i.e. cryosparc .cs file).
filename: /em_data/berk/cryoSPARC_Projects/CS-simulation-berk-01/J80/imported/007551211375608392020_particle1257_vacuum_finiteDose.mrc
filetype: 0
header_only: 0
idx_start: 285
idx_limit: 286
eer_upsampfactor: 2
eer_numfractions: 40
num_threads: 6
buffer: (nil)
buffer_sz: 0
nx, ny, nz: 0 0 0
dtype: 0
total_time: -1.000000
io_time: 0.000000
I created the .mrc files myself, and I am certain they don’t have any information regarding the eer_upsamplefactor or eer_numfractions. There is a mismatch, and I cannot really see why/how?
My .star file has the following format:
data_particles
loop_
_rlnImageName #1
_rlnMicrographName #2
_rlnCoordinateX #3
_rlnCoordinateY #4
01@particle0_vacuum_finiteDose.mrc particle0_vacuum_finiteDose.mrc 124 124
02@particle1000_vacuum_finiteDose.mrc particle1000_vacuum_finiteDose.mrc 124 124
03@particle1001_vacuum_finiteDose.mrc particle1001_vacuum_finiteDose.mrc 124 124
04@particle1002_vacuum_finiteDose.mrc particle1002_vacuum_finiteDose.mrc 124 124
...
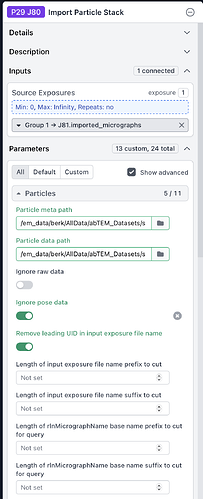
I’m also attaching images of my job configuration.
Has anyone managed to import simulated micrographs as particles into CryoSPARC before? Any advice or insights would be greatly appreciated.
Best regards,
Berk
(I couldn’t share the second screenshot as new users are only allowed one image)