Hi,
I tried to use the pyem package command of csparc2star.py to convert the .cs file of cryosparc into a relion 3.1 star file and didn’t work for me.
python csparc2star.py --copy-micrograph-coordinates particles.star particles_selected.cs PC2.star
Traceback (most recent call last):
File "csparc2star.py", line 28, in
from pyem import metadata
File "/home/greg/pyem/pyem/ **init** .py", line 3, in
from . import metadata
File "/home/greg/pyem/pyem/metadata.py", line 399
ptdf = util.dataframe_from_records_mapped(pt, {**general, **micrograph})
^
SyntaxError: invalid syntax
python --version
Python 2.7.15 :: Anaconda, Inc.
python -c 'import pandas; print pandas. **version** '
0.21.0
any idea about how to solve this problem ?
Hi @biolifei,
Where does your particles_selected.cs come from?
Typically, I used the cs file from 2D classification. For example, use command:
/Path_of_yours/csparc2star.py --swapxy cryosparc_P22_J220_020_particles.cs P22_J220_passthrough_particles.cs /Path_of_yours/from_P22_J220.star
swapxy seems necessary in my case. You can try to compare the max value of your coordinate between the star file generated by relion and converted cs file.
If you want reextract particles in Relion, then you may need to change the path in star file:
sed -i 's+J12/motioncorrected+MotionCorr/job006/Micrographs+g' from_P22_J220.star
and change the file name
sed -i 's+_patch_aligned_doseweighted.mrc+.mrc+g' from_P22_J220.star
those two steps trick relion to extract particles from file of its own MotionCorr mrc.
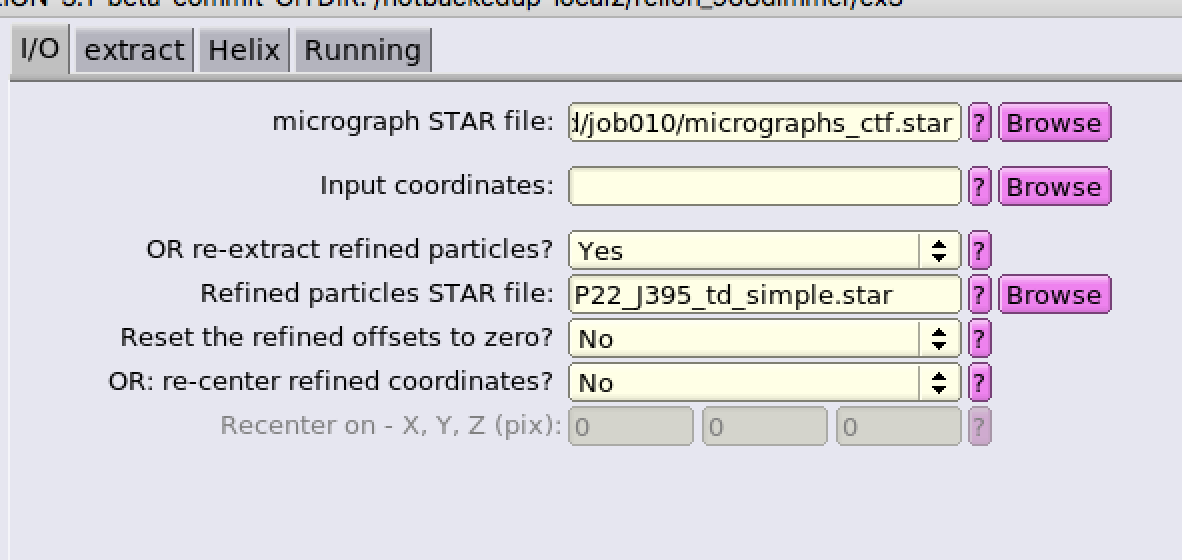
Then you may want to edit the metadata of star file like figure below (all the parameters should be in one row, rather than tow rows shown here):
And delete/edit some columns. In my case, I divided the coordinates by 2, and flip y coordinate using awk command by bash, and deleted defocus U/V column (I am not sure which columns are necessary, but seems like my star worked fine. Any suggestion)
Then using re-extract refined particles to extract
Seems like my re-extract particles are all not centered somehow. Any suggestion?
Best,
Feng10
@biolifei You must use Python 3. The project wiki has instructions on using conda to create a dedicated environment.
@Feng10 not sure what you mean. You mean the coordinates are completely wrong? If you have correct coordinates that you just want to re-center using refined offsets, you must tell Relion to do that.
Thanks for your reply. I tried to install the python with miniconda3. the installation is successful.
there is a problem for the follwing:
conda install -c conda-forge pyfftw healpy pathos
Solving environment: failed
PackagesNotFoundError: The following packages are not available from current channels:
- healpy
- pyfftw
- pathos
- multiprocess[version=‘>=0.70.4’]
- pathos
- ppft[version=‘>=1.6.4.5’]
Current channels:
To search for alternate channels that may provide the conda package you’re
looking for, navigate to
https://anaconda.org
and use the search bar at the top of the page.
so I installed the local files which downloaded from the website.
conda install --use-local pyfftw-0.12.0-py37hc3ef4f8_1.tar.bz2
Downloading and Extracting Packages
######################################################################## | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(pyem):~$ conda install --use-local healpy-1.13.0-py37hf476568_1.tar.bz2
Downloading and Extracting Packages
######################################################################## | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(pyem) :~$ conda install --use-local pathos-0.2.1-py36_1.tar.bz2
It shows that
Installing collected packages: pyem
Running setup.py develop for pyem
Successfully installed pyem
But when i ran the conversion command, i got the following messages:
python3.7 csparc2star.py --copy-micrograph-coordinates particles.star particles_selected.cs --loglevel debug PC2_del.star
Traceback (most recent call last):
File “csparc2star.py”, line 28, in
from pyem import metadata
File “/home/greg/pyem/pyem/init.py”, line 3, in
from . import metadata
File “/home/greg/pyem/pyem/metadata.py”, line 23, in
from . import star
File “/home/greg/pyem/pyem/star.py”, line 27, in
from pyem.geom import e2r_vec
File “/home/greg/pyem/pyem/geom/init.py”, line 4, in
from .geom import *
File “/home/greg/pyem/pyem/geom/geom.py”, line 20, in
from .quat_numba import distq
File “/home/greg/pyem/pyem/geom/quat_numba.py”, line 98, in
@numba.jit(cache=False, nopython=True, parallel=True)
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/decorators.py”, line 141, in jit
raise RuntimeError(msg)
RuntimeError: The ‘parallel’ target is not currently supported on Windows operating systems when using Python 2.7, or on 32 bit hardware.
Could you tell me any clue about how to solve this problem ?
Thanks in advance
RuntimeError: The ‘parallel’ target is not currently supported on Windows operating systems when using Python 2.7, or on 32 bit hardware.
I’m afraid you will need to address one of those issues; Numba is a hard dependency. You can also edit pyem/pyem/geom/quat_numba.py and set parallel=False and see how it goes. It may take several minutes instead of < 1 min to convert angles this way.
after I changed the pyem/pyem/geom/quat_numba.py and set parallel=False,
I have another probelm.
(pyem) :~/pyem$ python csparc2star.py --copy-micrograph-coordinates particles.star particles_selected.cs PC2_del.star
Traceback (most recent call last):
File “csparc2star.py”, line 28, in
from pyem import metadata
File “/home/greg/pyem/pyem/init.py”, line 3, in
from . import metadata
File “/home/greg/pyem/pyem/metadata.py”, line 23, in
from . import star
File “/home/greg/pyem/pyem/star.py”, line 29, in
from pyem.util import natsort_values
File “/home/greg/pyem/pyem/util/init.py”, line 3, in
from .util import *
File “/home/greg/pyem/pyem/util/util.py”, line 28, in
from … import vop
File “/home/greg/pyem/pyem/vop/init.py”, line 3, in
from .vop import *
File “/home/greg/pyem/pyem/vop/vop.py”, line 22, in
from pyfftw.interfaces.numpy_fft import rfftn
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/pyfftw/init.py”, line 18, in
from .pyfftw import (
ModuleNotFoundError: No module named ‘pyfftw.pyfftw’
here is another probelm after i changed the pyem/pyem/geom/quat_numba.py and set parallel=False ,
$ python csparc2star.py --copy-micrograph-coordinates particles.star particles_selected.cs PC2_test.star
Traceback (most recent call last):
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typeconv/typeconv.py”, line 4, in
from numba.core.typeconv import _typeconv
ImportError: cannot import name ‘_typeconv’ from ‘numba.core.typeconv’ (/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typeconv/init.py)
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File “csparc2star.py”, line 28, in
from pyem import metadata
File “/home/greg/pyem/pyem/init.py”, line 3, in
from . import metadata
File “/home/greg/pyem/pyem/metadata.py”, line 23, in
from . import star
File “/home/greg/pyem/pyem/star.py”, line 27, in
from pyem.geom import e2r_vec
File “/home/greg/pyem/pyem/geom/init.py”, line 3, in
from .convert_numba import *
File “/home/greg/pyem/pyem/geom/convert_numba.py”, line 18, in
import numba
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/init.py”, line 19, in
from numba.misc.special import (
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/misc/special.py”, line 3, in
from numba.core.typing.typeof import typeof
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typing/init.py”, line 1, in
from .context import BaseContext, Context
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typing/context.py”, line 11, in
from numba.core.typeconv import Conversion, rules
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typeconv/rules.py”, line 2, in
from .typeconv import TypeManager, TypeCastingRules
File “/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typeconv/typeconv.py”, line 17, in
raise ImportError(msg % (url, reportme, str(e), sys.executable))
ImportError: Numba could not be imported.
If you are seeing this message and are undertaking Numba development work, you may need to re-run:
python setup.py build_ext --inplace
(Also, please check the development set up guide http://numba.pydata.org/numba-doc/latest/developer/contributing.html.)
If you are not working on Numba development:
Please report the error message and traceback, along with a minimal reproducer
at: Sign in to GitHub · GitHub
If more help is needed please feel free to speak to the Numba core developers
directly at: https://gitter.im/numba/numba
Thanks in advance for your help in improving Numba!
The original error was: ‘cannot import name ‘_typeconv’ from ‘numba.core.typeconv’ (/home/greg/miniconda3/envs/pyem/lib/python3.7/site-packages/numba/core/typeconv/init.py)’
If possible please include the following in your error report:
sys.executable: /home/greg/miniconda3/envs/pyem/bin/python
Are you sure you’re using the correct python binary, with all the dependencies?
And you’re running 32-bit Windows on a very old computer?
python version under pyem environment is 3.7.x, i install the dependencies as the ones listed on the wiki.
The computer is GPUBox with 4x nvidia geforce gtx 1080 for the cryo-EM data processing with ubuntu 16.04 version
Something is not right, then. The instructions from the wiki should have worked verbatim. And there is certainly an appropriate build of pyfftw in conda-forge.
I suspect you are having an environment issue. What python do you have from which python and which -a python? My programs use #!/usr/bin/env python to run using the default environment python, when they are called as executables (and the conda env is activated). Explicitly using the python from your conda env will also work.
the problem is like here.
conda install -c conda-forge pyfftw healpy pathos
Solving environment: failed
PackagesNotFoundError: The following packages are not available from current channels:
healpy
pyfftw
pathos
multiprocess[version=’>=0.70.4’]
pathos
ppft[version=’>=1.6.4.5’]
Current channels:
https://conda.anaconda.org/conda-forge/linux-32
https://conda.anaconda.org/conda-forge/noarch
https://repo.anaconda.com/pkgs/main/linux-32
https://repo.anaconda.com/pkgs/main/noarch
https://repo.anaconda.com/pkgs/free/linux-32
https://repo.anaconda.com/pkgs/free/noarch
https://repo.anaconda.com/pkgs/r/linux-32
https://repo.anaconda.com/pkgs/r/noarch
https://repo.anaconda.com/pkgs/pro/linux-32
https://repo.anaconda.com/pkgs/pro/noarch
To search for alternate channels that may provide the conda package you’re
looking for, navigate to
https://anaconda.org
and use the search bar at the top of the page.
so I installed the local files which downloaded from the website.
conda install --use-local pyfftw-0.12.0-py37hc3ef4f8_1.tar.bz2
Downloading and Extracting Packages
######################################################################## | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(pyem):~$ conda install --use-local healpy-1.13.0-py37hf476568_1.tar.bz2
Downloading and Extracting Packages
######################################################################## | 100%
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
(pyem) :~$ conda install --use-local pathos-0.2.1-py36_1.tar.bz2
Wait, are you actually using 32-bit Linux? It looks like you only have 32-bit conda channels. Please post:
uname -a
lsb_release -a
Thanks a lot for your help. here is the information you asked.
(pyem) ~/pyem$ conda activate pyem
(pyem) :~/pyem$ which python
/home/greg/miniconda3/envs/pyem/bin/python
(pyem) :~/pyem$ which -a python
/home/greg/miniconda3/envs/pyem/bin/python
/home/greg/miniconda3/bin/python
/usr/bin/python
~/pyem$ uname -a
Linux pcbi00238 4.4.0-179-generic #209-Ubuntu SMP Fri Apr 24 17:48:44 UTC 2020 x86_64 x86_64 x86_64 GNU/Linux
(pyem) :~/pyem$ lsb_release -a
LSB Version: core-9.20160110ubuntu0.2-amd64:core-9.20160110ubuntu0.2-noarch:printing-9.20160110ubuntu0.2-amd64:printing-9.20160110ubuntu0.2-noarch:security-9.20160110ubuntu0.2-amd64:security-9.20160110ubuntu0.2-noarch
Distributor ID: Ubuntu
Description: Ubuntu 16.04.6 LTS
Release: 16.04
Codename: xenial
Which miniconda installer did you use?
Should be this 64-bit one, not the 32-bit one:
https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh
Daniel, Thanks a lot for your help!!!
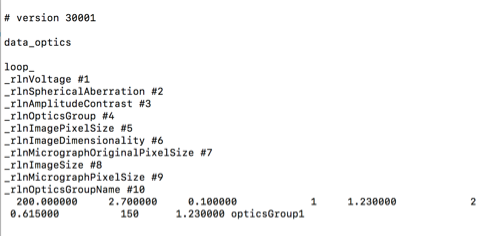
now I can re-extract the particles again from relion, after adding some parameters for the data optics
such as
version 30001
data_optics
loop_
_rlnOpticsGroupName #1
_rlnOpticsGroup #2
_rlnMtfFileName #3
_rlnMicrographOriginalPixelSize #4
_rlnVoltage #5
_rlnSphericalAberration #6
_rlnAmplitudeContrast #7
_rlnImagePixelSize #8
_rlnImageSize #9
_rlnImageDimensionality #10
opticsGroup1 1



Don’t forget to revert your modified pyem files, if you are still using them.
git reset --hard origin/master should do it.