Hi all,
I have processed a model to 3.10 Ang using movies obtained from Glacios-TEM equipped with a Falcon IV 4K at a pixel size of 1.5 Ang/pixel. Given the diagnostic cFSCs plot shown during NU-refinement, much of my signal is at the Nyquist limit, so I decided to EER upsample my original movies twofold to 0.75 Ang/pixel to squeeze out as much resolution as possible.
I have performed new patch motion/CTF jobs as it seems that motion data from the 1.5 A/pix data cannot be retained. However, I am wondering if I can import the RBMC hyperparameters from the 1.5 A/pix data to my upsampled 0.75 A/pix data, or perform a new RBMC job? If this does not work, please inform me the reasons why, as well as general tips/expectations for upsampling!
Thank you so much.
I’d certainly try just manually setting the various hyperparameters (from the logged output of the previous run) and see what the result is - it would be an interesting exercise which could help the rest of the community! 
If you then run a fresh estimation, it would also be interesting to compare them!
Thank you! I’ll keep you posted on the results.
1 Like
Pre motion correction:
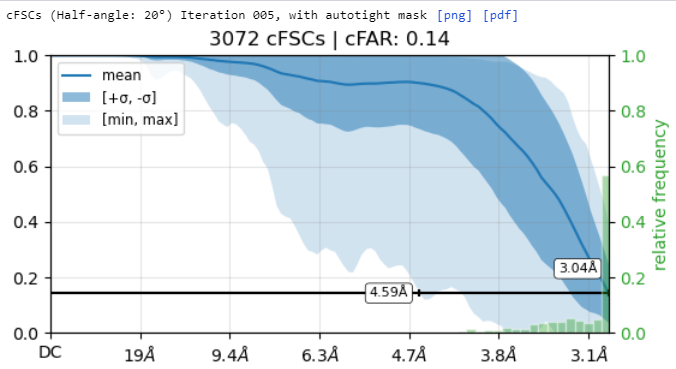
Res of 3.21 Ang
Old values:
Spatial prior strength: 6.4276e-03
Spatial correlation distance: 500
Acceleration prior strength: 1.9641e-03
Res of 3.10 Ang
cFSCs show signals ranging from 3.86 - 2.80 Ang
New values:
Spatial prior strength: 6.3103e-03
Spatial correlation distance: 500
Acceleration prior strength: 3.7268e-02
Res of 3.07 Ang
cFSCs show signals ranging from 3.09 to 2.78 Ang
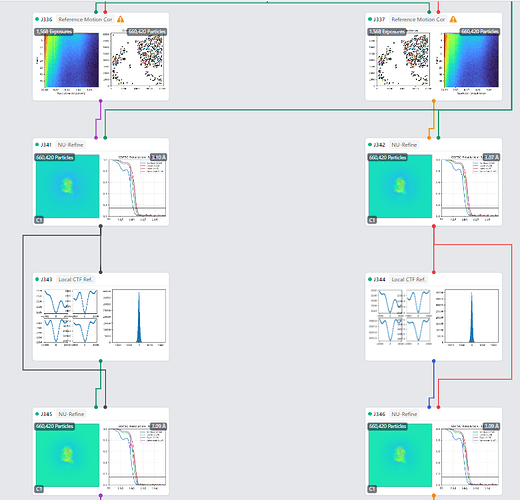
An image of the workflow because I can only have one photo in a single post. How much should I trust the estimated resolution? Also, after local CTF correction, the res seems to both be 3.09 Ang? Would this mean the difference is negligible?
Yes.  That’s good, although while the acceleration prior is an order of magnitude different (well, half an order) the spatials show consistent results so I’d go with it.
That’s good, although while the acceleration prior is an order of magnitude different (well, half an order) the spatials show consistent results so I’d go with it.
That said, are the cFSCs really that broad for the old values? I’m a little surprised that they’re so much broader than the new values. If you do a 3D classification, do you end up with much in lower res classes?
To be honest, a difference of resolution of 10% when >3A doesn’t make much difference. It’s impossible to visually distinguish the difference between a “3A” map and a “3.1A” map.  Building a model into a “3A” map is pretty much the same as building into a “3.3A” map. It only starts to make much difference when you can start seeing smaller sidechains, holes in aromatic rings, etc…
Building a model into a “3A” map is pretty much the same as building into a “3.3A” map. It only starts to make much difference when you can start seeing smaller sidechains, holes in aromatic rings, etc…
Apologies for my typo, with old values it is 3.86 to 2.80 Ang, and new values it is actually 4.09 to 2.78 Ang. However, as you’ve said, this much difference won’t affect model building.
Within this dataset, 3D classifications have usually yielded slightly lower resolutions without meaningful conformational differences, which is why I’ve not had to separate into classes. Unfortunately, I’ve cleared the jobs to make space as we were running out…
Thank you for your help!
Ah, typo. Got it. No worries.
If you get very similar 3D classes, set class similarity to 0, or 0.00001… it’s the only way I’ve been able to clean some junk out - I think alignment off 3D classification in RELION does a better job without fiddling around, but find the right settings in CryoSPARC 3D class and it does pretty well too. 
2 Likes