I’ve run a Topaz denoise job with its pre-trained model and it did make my particles slightly easier to see. Given that all of my micrographs have 1/5-1/4 of the area covered by carbon (this was intentional as my particles tend to appear at the edges of the hole), I was wondering if the carbon would negatively impact Topaz denoise. I assume the answer is yes as Topaz normalizes the micrographs and too much carbon would skew the intensity distribution. Is this assumption correct? I’m considering segmenting out carbon, replacing it with the mean intensity value in the non-carbon area, and Topaz denoise again. Would this be a reasonable thing to do? Any insights would be appreciated!
Hi csparc_addict, please pardon my not having a direct technical answer, but I do want to mention that I have used Topaz and crYOLO with samples on continuous amorphous carbon without denoising, and they work quite well. I can understand if you still want to improve visibility, but I just wanted to mention this in case it saves you unnecessary work.
I would try the (trained on your data) cryosparc denoising approach (Micrograph Denoise). It gives much better results than topaz denoise in my hands.
Agreed, with one additional point - train the model yourself on that dataset. Doesn’t take very long, and quality of results can change significantly. I’ve posted examples previously of how much difference it can make, but as I can’t find the thread right now…
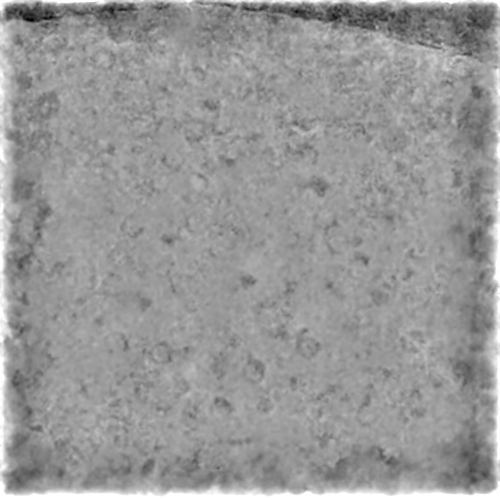
Pre-trained:
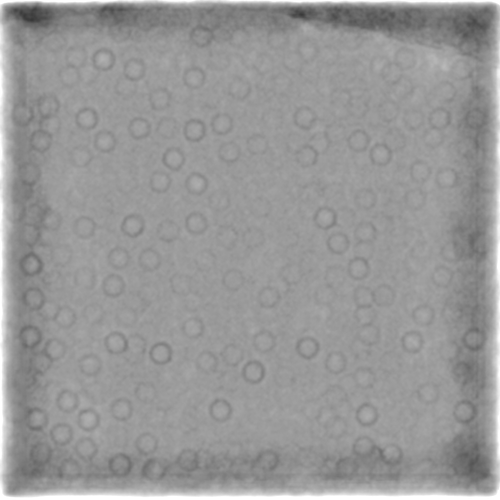
Self-trained:
(Same micrograph)
yes absolutely, trained on your data was what I was referring to (and the key difference with topaz denoise I suspect)
I thought you were, but thought it worth stating explicitly. ![]()
Hi drichman, thanks for your input. Actually using Topaz to pick on the un-denoised micrographs was the first thing I tried. Unfortunately it didn’t work well. I guess it’s really a case-by-case situation.
Hi @olibclarke , @rbs_sci ,
I followed your suggestions and used cryosparc’s denoising approach with a self-trained model to denoise my micrographs. Consistent with your observations, it worked a lot better than topaz denoise. At this point I’m curious about the underlying principle of the approach and how it differs from topaz denoise. It seems like another deep-learning method but is it not based on the noise2noise framework?
One thing I don’t like about the denoised micrographs is the contrast of the carbon gets greatly enhanced and the edges are extremely bright. The job parameter “Greyscale normalization factor” indicates that intensity normalization is still applied. If this is true, wouldn’t the concern that too much carbon would skew the intensity distribution still hold?
Thanks a lot for your help!
It is based on noise2noise - the difference as I understand it lies in the training, which @rwaldo has explained in detail in another post: Applying micrograph denoiser to motion corrected data - query - #4 by rposert
I actually like this (mostly) as it makes removing carbon picks and edge picks a lot easier - less loss of actual particles, particularly on low defocus micrographs! ![]()