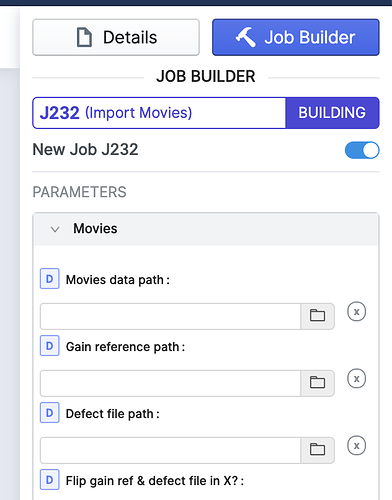
Is there any way to incorporate defect file (.mrc) in cryosparc while importing movies?
HI @Anshul, please specify the defect file path in the Job Builder for Import Movies:
Hi @Anshul - the defects file should be a text file, e.g. as generated by DM, not an mrc file.
Hi
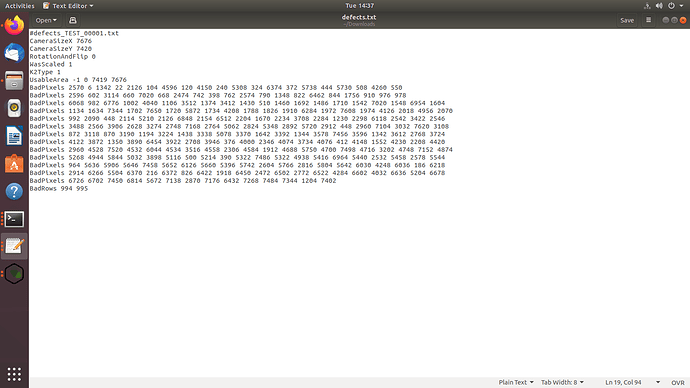
Can we convert this defect file in mrc format into a text (.txt) file
I don’t know, but I am assuming the mrc file was created from a defects.txt file, as described here (https://bio3d.colorado.edu/SerialEM/hlp/html/about_camera.htm) - perhaps you can obtain it from whoever collected the data?
Hi @olibclarke @spunjani ,
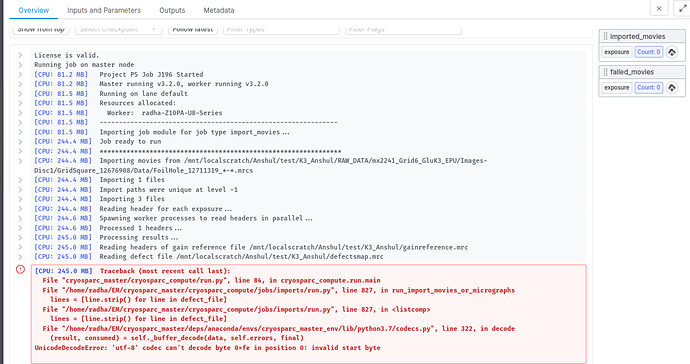
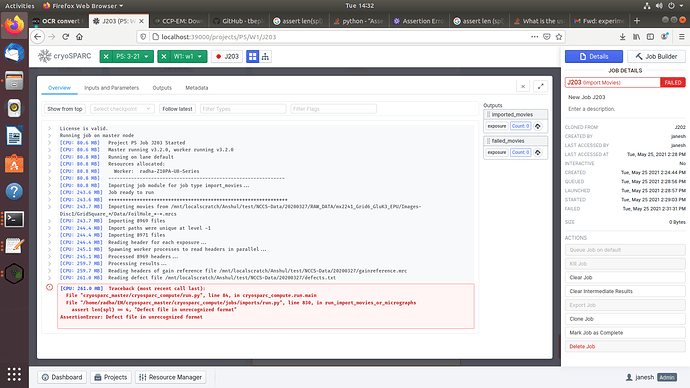
As per your suggestions, I got the defect file in text format and tried to import the movies again but still the job is failing and showing this error. I am attaching the defect file and snapshot of the error.
Hi @olibclarke @spunjani,
this is the error message
Hi @Anshul,
Unfortunately there are several different defect file text formats, and not all of them are supported yet. There is a workaround, however: any pixels that have a value of zero in the gain reference file are treated as defects. So, if you have a defect file in mrc format, verify that it has ones on good pixels and zeros on defective pixels, and then multiply that in to your actual gain reference file.
–Harris
alternatively, if you have cisTEM handy, just run remove_outlier_pixels on your gain reference - we do this when we have obvious outlier pixels and it does the trick
I encountered the same problem, with a large stripe on all movies and a defect text file not recognized by cryoSPARC (v.3.2.0).
The defect zone has pixel values of 1 in the gain.mrc file. I tried the remove_outlier_pixels from cisTEM, but it didn’t work. So I tried to find a way to change these defective pixel values to 0 in the gain file. I managed to do this with three IMOD commands:
1. Generate a defect file in mrc format
clip defect -D defects.txt fileWithFrames defects.mrc
where the fileWithFrames is used only to set the size of the output and can be any file of the right X and Y size (clip is only available in IMOD 4.10.7 or higher).
This will generate a mrc file with a pixel value of 1 for defective pixels, and 0 for the others.
2. Subtract the defect.mrc from the gain.mrc
subimage gain.mrc defects.mrc gain_subimage.mrc
This will generate a gain_subimage.mrc with a pixel value of zero for defective pixels and unchanged values for the other pixels in the gain file. (The command clip subtract does not work because the gain.mrc is a mode 2 map, and the defect.mrc is a mode 0).
3. Correct the pixel size in the gain_subimage.mrc file
After subtraction the pixel size is messed up because it was different in the gain.mrc file (pixel size 1) and the defect.mrc (pixel size depending on camera).
alterheader -fixpixel gain_subimage.mrc
will reset the pixel size to 1 in the gain_subimage.mrc.
Now you should have a proper gain file with a pixel value of 0 for defective pixels that will be recognized as such by cryoSPARC. It worked in my case.
What exactly do you mean with fileWithFrames? what is the input for that?
Any plans to make the missing ones supported by cryoSPARC then?
-Andre