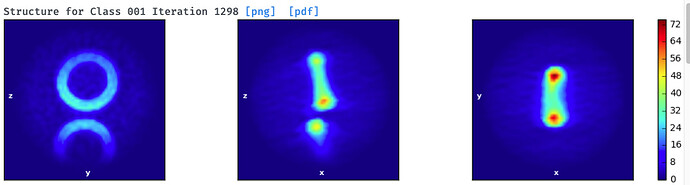
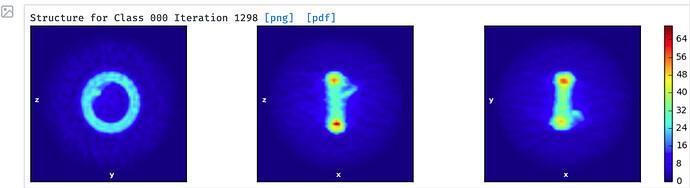
I am working with a highly symmetrical circular particle. Both blob and template picking work fine. But I have difficulty get rid of neighboring particles. After several rounds of 2D classification and Select 2D for single particle classes (top panel), ab-initial model building still put majority of the particles to model with neighbors (middle panel) instead of single particle model (bottom panel).
Hi @Ricky,
In this case, do you only see top views of the circle, as your 2D class image suggests?
One idea might be to re-pick the particles using the best 2D templates you have, which will encourage the picks to be more centered.
Also, it may be worth simply using all the particles and the single-molecule initial model from ab-initio to perform a refinement (without symmetry at first). The double-particle class seems to be composed of otherwise good particles, which will naturally align with the single-particle class if you use that as an initial model for refinement. You can then use heterogeneous refinement to see any heterogeneity after the homogeneous refinement, and repeat the refinement with symmetry.
In other words, the particles do not all have to go into the same ab-initio class in order to be all used in the same refinement.
Let us know what you figure out!
Some other thoughts:
- Re-make the grid at lower particle density to avoid the neighboring particles. Will require collecting more images, obviously.
- Have you considered that your structure actually dimerizes in some way that may be biologically relevant? Re-extract the particles with a slightly bigger box, intentionally select the apparent dimers, and reconstruct them to analyse their interface.
- 3D refine the dimer class (to align them) then do particle subtraction and local refinement of one the rings. Tutorial on this here: https://cryosparc.com/blog/local-refinement-snRNP-case-study/. This at least lets you use 1 particle in each pair.
3a. Modification of #3: You might be able to use both particles in the pair by subtracting either one into two separate particle stacks, doing two local refinements, then combining those into a single refinement. That’s an idea, not something I know will work for sure. YMMV. - I notice that your single paricle structure has a little blip sticking out of it which does not appear in the dimer structure. This blip may (or may not) be relevant. Just something that catches my eye.