Hi all,
I am learning 3D variability analysis in cryoSPARC from the tutorials Part One and Part Two. I am currently trying on the EMPIAR10076 dataset.
In my understanding, I need to obtain a consensus reconstruction before the 3D variability, which can be achieved by
131,899 particles were processed, first through ab-initio reconstruction (single class) then homogeneous refinement to obtain a consensus refinement of the ribosome. This structure showed regions of low and high resolution where there was variability.
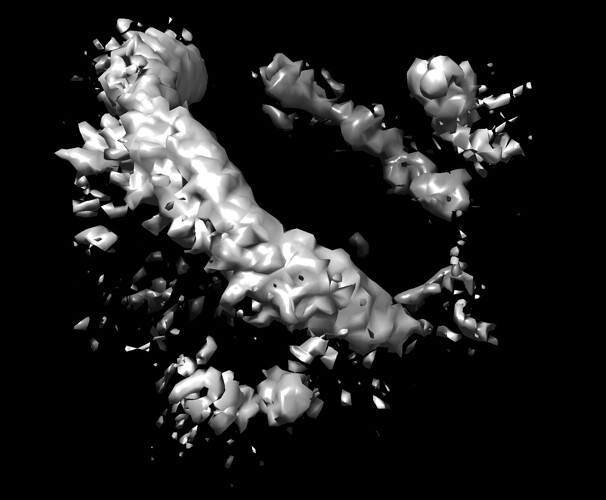
However, after loading all the particle images via the .star file, I can only get such structures from my trials:
I see the inner of the structure is almost empty.
Could you suggest what a reasonable structure should look like, and what resolution it can achieve? Also, any tips (e.g. on parameter setting) to get the structure are welcome. Thanks!