Hello Everyone
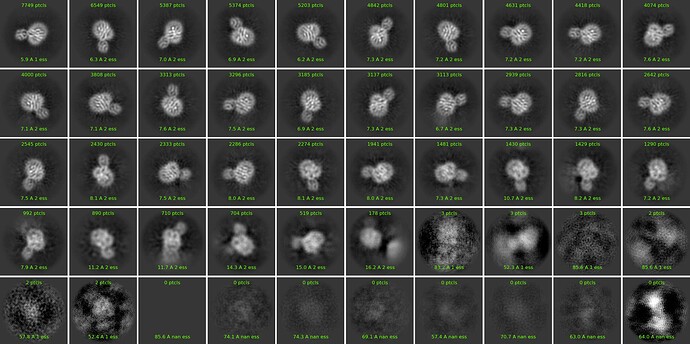
I have a dataset for a small membrane protein in detergent. I could see good 2D classes with transmembrane domains, however, I am unable to get a good 3D reconstruction. Maps are all at7-8 Ao. I am trying to get a high-resolution map. I thought if we are seeing good 2Ds it means we have a rigid conformation and the software is able to align the particles well resulting in good 3D models. Am I wrong or are there any tricks that I have to use? Currently, I am using the default parameters. There are 108000 particles for this 2D.
Hi,
Maybe there are few viewing directions. Check your first round of 2D classification if there were some ‘bad’ classes of different viewing directions.
This looks promising, however you need to have two or better three times the number of particles you currently have; so acquire another large dataset, and merge the particles with this one. Your classes will improve (they’re a bit fuzzy now). How big is this protein, probably 70-90 kDa ? Maybe, then you will be able to get a decent ab-initio. Cheers
Looks to me like an orientation bias issue - these all look like top or slightly oblique views, no clear side views.
I would experiment with adding a small amount of high CMC detergent (CHAPS, CHAPSO, fluorinated foscholine), using a thin carbon or graphene substrate, or collecting with a tilt (more or less in that order of preference.
Hello @marino-j my protein is 70kd. Thank you for the suggestion. I will try to collect another larger dataset.
Thank You @olibclarke. I can try adding chaps to see if it can improve the preferred orientation problem. Also, I only get ~100000 particles with good 2D from~ 3M particles coming from ~3000 micrographs.
Thank You @Lai I tried adding some classes back, but that did not improve my 3D models. Will repeat and see.
I’ve seen similar 2D classes that were only detergent. Microscopes and cameras are so good that we can now do structures of detergent micelles! I hope it’s not the case for you (you indeed have a side density that acts in your favor), but keep it in mind in case you can’t go any further.
Good luck
Vincent