Hi,
We run Live on core facility GPUs during data collection and find it extremely helpful for on the fly decision making and assessment of data quality.
In particular, the “streaming” exposure curation tools are wonderful, and greatly superior to those currently available in “regular” cryosparc.
We would love to be able to easily get a list of all good (accepted) micrographs after the completion of a Live session, so we can copy these and only these to our local GPU to restart processing (either in cryosparc or other packages)
Is there an easy way to do this currently?
Also it would be useful if these curation thresholds could still be tweaked after the completion of a session (we can only use Live during our Krios session, but being able to tweak curation thresholds afterwards would be useful before export/transfer).
Cheers
Oli
Hi @olibclarke,
If I understand correctly this should already be possible; you can choose to export exposures or particles from a Live session at any time: https://guide.cryosparc.com/live/new-live-session-start-to-finish-guide#export-exposures-and-or-particles
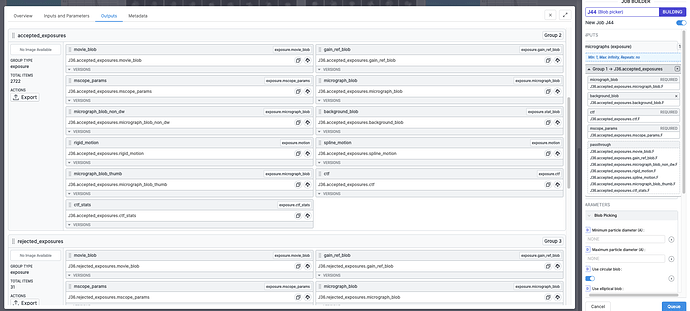
Here’s a screenshot of the accepted exposures (exposure exports contain four groups: all, accepted, rejected and manually rejected) being applied to a blob picker job:
There’s no need to redo motion correction or CTF estimation.
As per your request to export with custom thresholds applied, we can certainly add that to our to-do list.
- Suhail
Hi Suhail
That works fine for exporting to process in a cryosparc instance on the same workstation.
I would like to export for processing on a different workstation, with our copy of cryosparc. Is there an easy way to do that? I’m probably missing something obvious, sorry
I think this will probably be a pretty commonly requested workflow for users of core facilities - process data on the fly in csparc, then export the raw data corresponding to the good movies/mics/particles for further processing in cryosparc on a machine at their home institution.
Cheers
Oli
1 Like
Also there are reasons to redo motion correction etc - eg if we want to perform bayesian polishing in relion, we need to perform motion correction in relion. This sometimes still gives a boost in resolution and map quality compared to patch motion plus local motion in csparc.
@sdawood is there any easy way to do this? I can’t see anything on the command line where I can get a list of accepted exposures from the Live session directory, and when I export the exposures and try converting any of the various cs files for the accepted exposures to star none of them have the micrograph name preserved
Hi @olibclarke,
In the outputs tab of the regular cryoSPARC interface, you should be able to download a .cs file from the movie_blob output. This .cs file contains a movie_blob/path field, which I believe is what you’re looking for. You can follow this tutorial if you want to write a script in Python to read and extract this field from the .cs file: https://guide.cryosparc.com/processing-data/tutorials-and-case-studies/manipulating-.cs-files-created-by-cryosparc
- Suhail
Great I will have a go at this! It would be good if this was easier though - I think transferring raw/processed data from a facility GPU to a local GPU is a pretty common workflow
For sure, we’ll evaluate options for making this data easier to retrieve!
not just this metadata - exporting the session including raw data, to make transfer to either another computer on the network or an external hard drive easier, for easier import into another instance of cryosparc on a different workstation
2 Likes
@DanielAsarnow is there any way to do this using csparc2star.py?
If I run csparc2star.py like so:
csparc2star.py P5_S15_accepted_live_exposures.cs P5_S15_accepted_live_exposures.star
The resulting star file doesn’t have any micrograph or movie names - is there a way to make it keep them intact?
data_optics
loop_
_rlnVoltage #1
_rlnSphericalAberration #2
_rlnAmplitudeContrast #3
_rlnOpticsGroup #4
_rlnImageDimensionality #5
300.000000 2.700000 0.100000 2 2
data_micrographs
loop_
_rlnDefocusU #1
_rlnDefocusV #2
_rlnDefocusAngle #3
_rlnPhaseShift #4
_rlnCtfMaxResolution #5
_rlnCtfFigureOfMerit #6
_rlnOpticsGroup #7
21325.050781 21257.533203 16.890060 0.000000 2.857556 0.000000 2
25198.642578 25005.318359 -1.398781 0.000000 3.123030 0.000000 2
27196.177734 26878.787109 14.257236 0.000000 3.175358 0.000000 2
23136.046875 22991.398438 16.272697 0.000000 3.500672 0.000000 2
25924.529297 25706.460938 15.474290 0.000000 3.113364 0.000000 2
24666.857422 24481.470703 9.189087 0.000000 3.142540 0.000000 2
27473.302734 27182.570312 14.404624 0.000000 3.142193 0.000000 2
23432.986328 23292.095703 11.442520 0.000000 3.229946 0.000000 2
26245.246094 25999.320312 11.835846 0.000000 3.099082 0.000000 2
1 Like
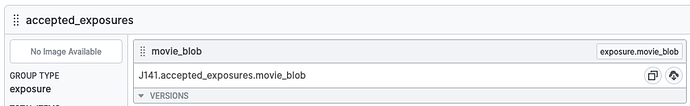
I found an easier way to do this - export a result group (from a completed Live Exposure Export Job, in the “outputs” tab), e.g. accepted exposures (button on left):
This then creates a new exported folder that has links to the accepted motion corrected mics in a sub directory, e.g. /P5/exports/groups/P5_J291_accepted_exposures/S15/motioncorrected
I can then make a text file with a list of these and use them to filter the micrographs/movies either during or after transfer. A little convoluted, but it works!
9 Likes
Hi @olibclarke!
Did you ever get a reply from @DanielAsarnow ?
Do you filter micrographs on-the-fly data transfer by submitting that text file directly to the transfer software in the command? Or do you have any bash/python scripts? Such scripts should not be that difficult to figure out, but would you like to share any script or experience in this data transfer step? 
I guess it could be useful for the whole community to have nice procedures to clear the clutter… data storage is becoming a problem everywhere. Most people do not know how to clearly sort things, some do not dare, others do not care… but some do 
Cheers,
André
I think the missing micrograph names can be added from the all exposures .cs file? You can also make the micrograph star for just the selected particles using star.py --to-micrographs which might be helpful for scripting something.
2 Likes