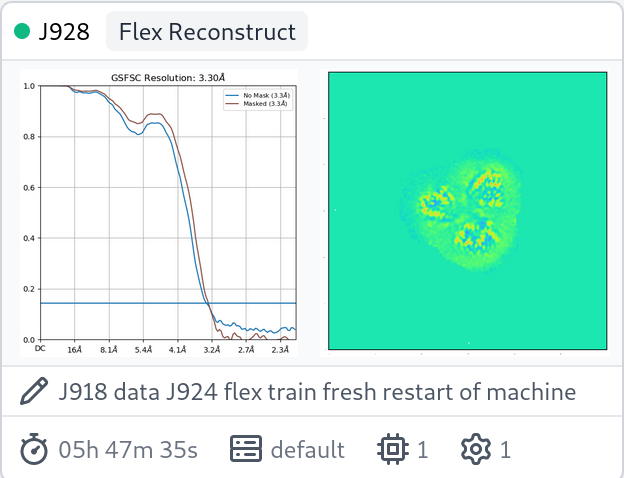
For one of my datasets, 3dflex reconstruct shows an odd rise in FSC. This is a C1 membrane protein. There are not a significant number of duplicate particles, if any.
FSC from 3dflex:
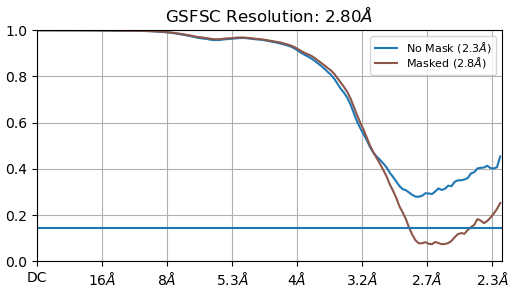
FSC from Non-uniform:
For one of my datasets, 3dflex reconstruct shows an odd rise in FSC. This is a C1 membrane protein. There are not a significant number of duplicate particles, if any.
FSC from 3dflex:

FSC from Non-uniform:

Hi @DanielAsarnow,
Thanks for the question. Seeing rises in the high-res portion of the FSC during 3DFlex Reconstruction is likely symptomatic of inadequate convergence of the optimization algorithm (L-BFGS) used during the reconstruction process. Since the half-maps are initialized to all zeros before reconstructing, we hypothesis that inadequate convergence manifests itself this way because BFGS focuses on optimizing low-frequency “directions” before high-frequency ones, thus the high-frequency portion of the half maps are close to identical. See the Parameter tuning section of the 3D Flex tutorial. The suggested fix is to significantly increase the number of Max BFGS iterations, perhaps to at least 40 or even 60, and re-run the reconstruction.
Best,
Michael
More iterations? Ouch…I hope the caching update is ready soon ![]()
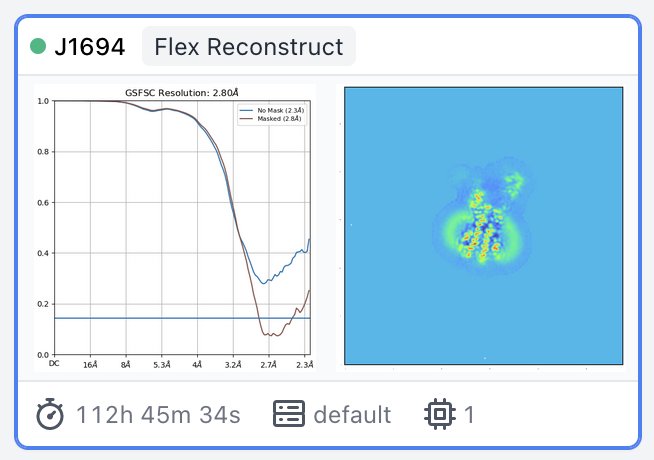
(112 hours for 1.2E6 particles at 144 px)
I had a similar issue with detergent solubilized membrane protein and found that 40 iterations was better than 20, and 60 better than 40. But, the unmasked FSC curve still fails to reach zero.
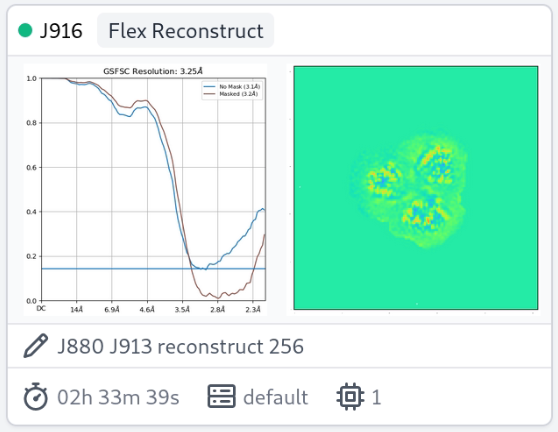
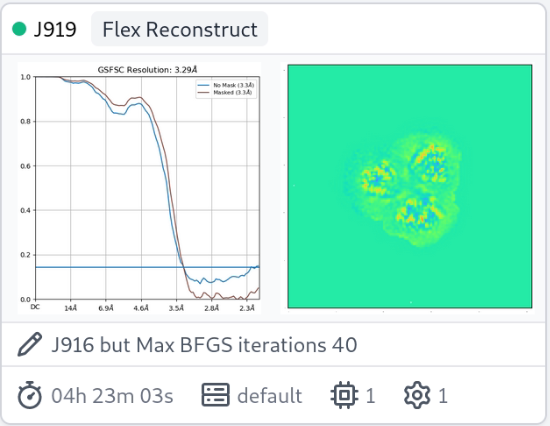
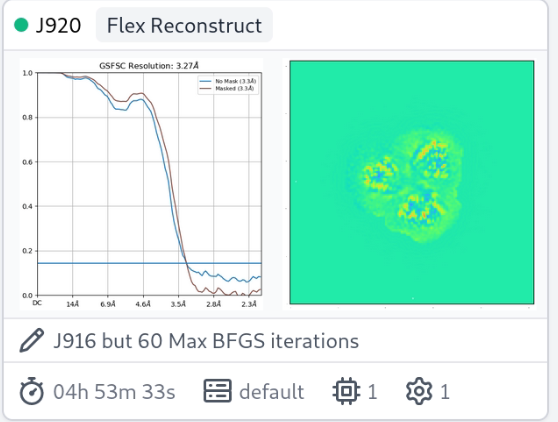
Nice, thanks for these! How many particles are there? The jobs are fast.
It looks like the spurious correlations are outside the mask, no?
Hi Daniel,
163,000 particles. I’m newish to cryoEM and membrane proteins and not sure about your comment regarding the mask - are you suggesting that a tighter mask would do better? More explanation would be helpful to me. My computer is home built with 2 intel‐i7 CPUs, 256‐GB RAM, 4 RTX2080 GPUs, 1 p600 GPU, 4 TB SSD scratch and 96 TB HD space configured as RAID.
Would it be good to try to improve further so the unmasked FSC curve does go to zero? I’m running 100 iterations now, and also trying to increase box/pix from 256/128 to 300/128 (300 is the box size I used in NU with per particle ctf and that gave 3.0 A resolution with a spike in the corrected FSC that looks nearly identical to what you originally posted).
I made the mask using UCSF Chimera by fitting the current model into the map created in Data Prep and then making a molmap. I then showed all atoms in spacefill and adjusted the threshold to contain the spacefill atoms plus a little more volume. Then I used vop threshold #0 minimum 0.03 set 0 to create map #1, followed by vop threshold #1 maximum 0.029 setMaximum 1 to create map #2, followed by vop resample #2 ongrid #0 to create the initial mask. I then imported that mask into cryosparc and using Vol Tools with 0.9 threshold padded it with 7 pixels. The jobs look like the following.
Thanks for you posting - it really fits my situation!
Tracy
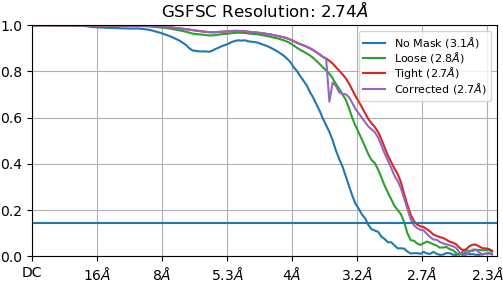
Turns out that 100 BFGS iterations was no different from 60, but increasing the box from 256 to 300 did do better. The first time I tried to run it the memory was not sufficient so it crashed. But, a fresh restart of my computer removed things from RAM and it was able to run consuming 190 GB of RAM. The first image is the Flex reconstruction with the bigger box, and the second image is the Validation FSC with a new mask generated by the utility.